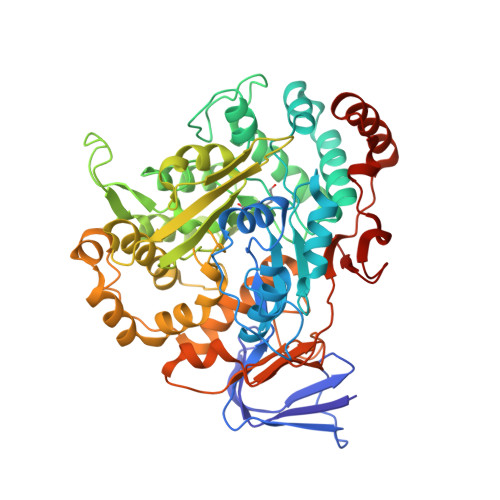
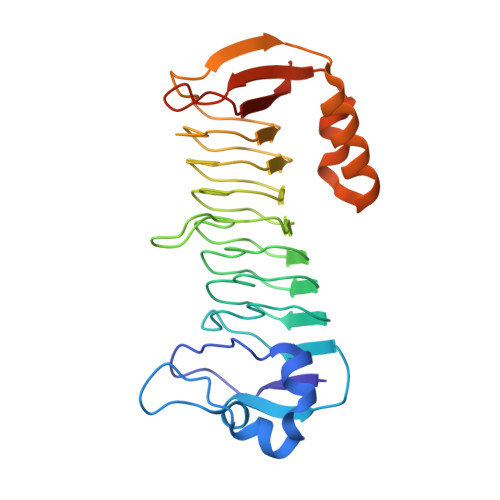
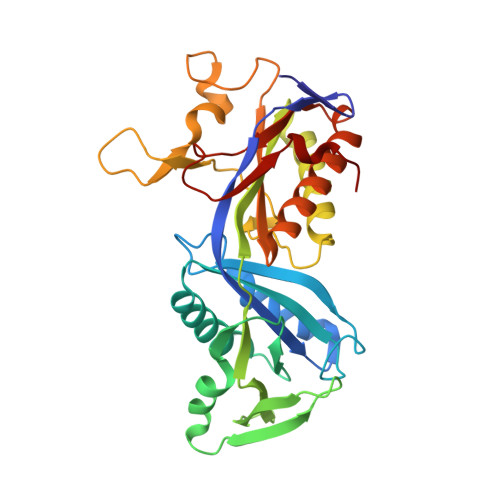
Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites.
Hemmann, J.L., Wagner, T., Shima, S., Vorholt, J.A.(2019) Proc Natl Acad Sci U S A 116: 25583-25590
- PubMed: 31776258
- DOI: https://doi.org/10.1073/pnas.1911595116
- Primary Citation of Related Structures:
6S6Y - PubMed Abstract:
Methylotrophy, the ability of microorganisms to grow on reduced one-carbon substrates such as methane or methanol, is a feature of various bacterial species. The prevailing oxidation pathway depends on tetrahydromethanopterin (H 4 MPT) and methylofuran (MYFR), an analog of methanofuran from methanogenic archaea. Formyltransferase/hydrolase complex (Fhc) generates formate from formyl-H 4 MPT in two consecutive reactions where MYFR acts as a carrier of one-carbon units. Recently, we chemically characterized MYFR from the model methylotroph Methylorubrum extorquens and identified an unusually long polyglutamate side chain of up to 24 glutamates. Here, we report on the crystal structure of Fhc to investigate the function of the polyglutamate side chain in MYFR and the relatedness of the enzyme complex with the orthologous enzymes in archaea. We identified MYFR as a prosthetic group that is tightly, but noncovalently, bound to Fhc. Surprisingly, the structure of Fhc together with MYFR revealed that the polyglutamate side chain of MYFR is branched and contains glutamates with amide bonds at both their α- and γ-carboxyl groups. This negatively charged and branched polyglutamate side chain interacts with a cluster of conserved positively charged residues of Fhc, allowing for strong interactions. The MYFR binding site is located equidistantly from the active site of the formyltransferase (FhcD) and metallo-hydrolase (FhcA). The polyglutamate serves therefore an additional function as a swinging linker to shuttle the one-carbon carrying amine between the two active sites, thereby likely increasing overall catalysis while decreasing the need for high intracellular MYFR concentrations.
- Institute of Microbiology, Eidgenössische Technische Hochschule Zurich, 8093 Zurich, Switzerland.
Organizational Affiliation: