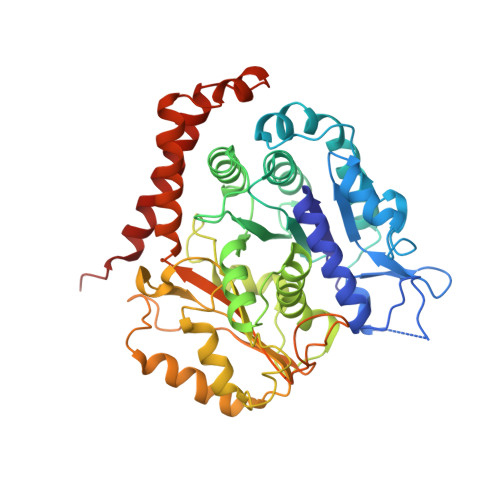
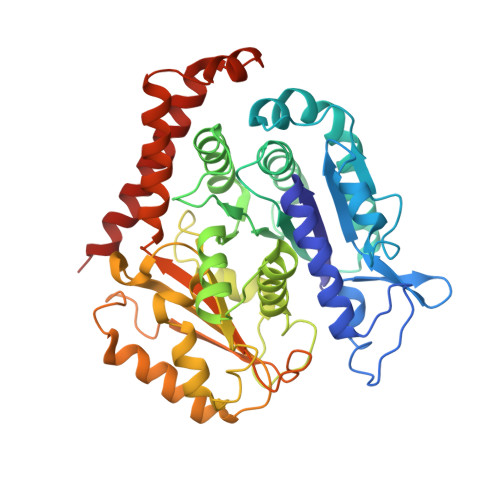
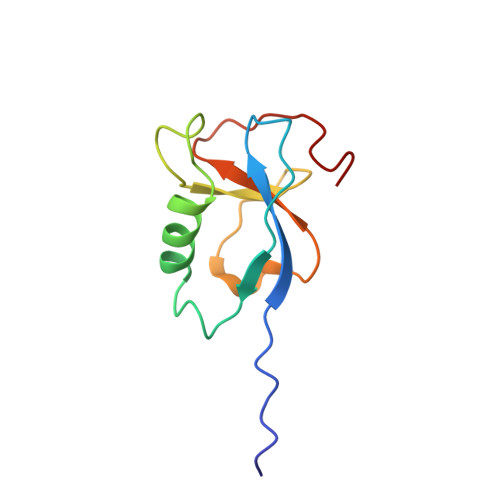
Pseudo-repeats in doublecortin make distinct mechanistic contributions to microtubule regulation.
Manka, S.W., Moores, C.A.(2020) EMBO Rep 21: e51534-e51534
- PubMed: 33051979
- DOI: https://doi.org/10.15252/embr.202051534
- Primary Citation of Related Structures:
6REV, 6RF2, 6RFD - PubMed Abstract:
Doublecortin (DCX) is a neuronal microtubule-associated protein (MAP) indispensable for brain development. Its flexibly linked doublecortin (DC) domains-NDC and CDC-mediate microtubule (MT) nucleation and stabilization, but it is unclear how. Using high-resolution time-resolved cryo-EM, we mapped NDC and CDC interactions with tubulin at different MT polymerization stages and studied their functional effects on MT dynamics using TIRF microscopy. Although coupled, each DC repeat within DCX appears to have a distinct role in MT nucleation and stabilization: CDC is a conformationally plastic module that appears to facilitate MT nucleation and stabilize tubulin-tubulin contacts in the nascent MT lattice, while NDC appears to be favored along the mature lattice, providing MT stabilization. Our structures of MT-bound DC domains also explain in unprecedented detail the DCX mutation-related brain defects observed in the clinic. This modular composition of DCX reflects a common design principle among MAPs where pseudo-repeats of tubulin/MT binding elements chaperone or stabilize distinct conformational transitions to regulate distinct stages of MT dynamic instability.
- Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck, University of London, London, UK.
Organizational Affiliation: