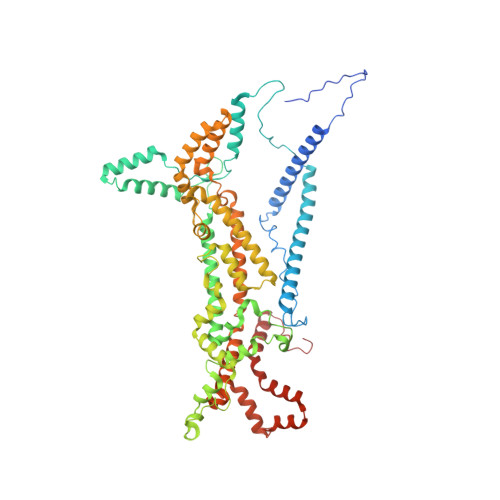
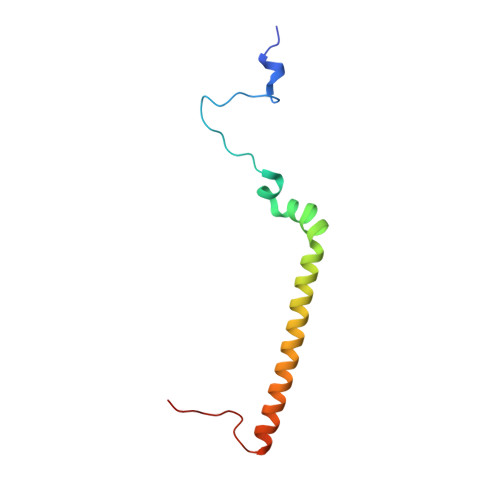
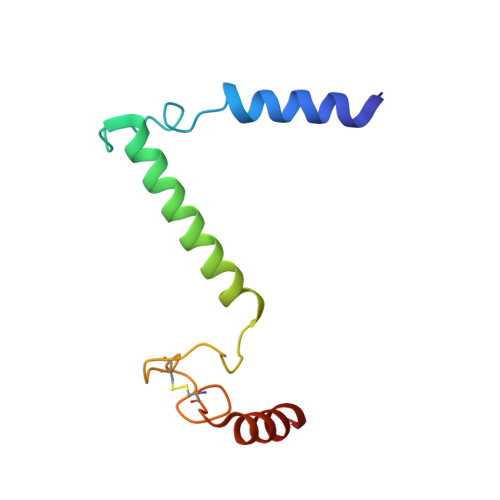
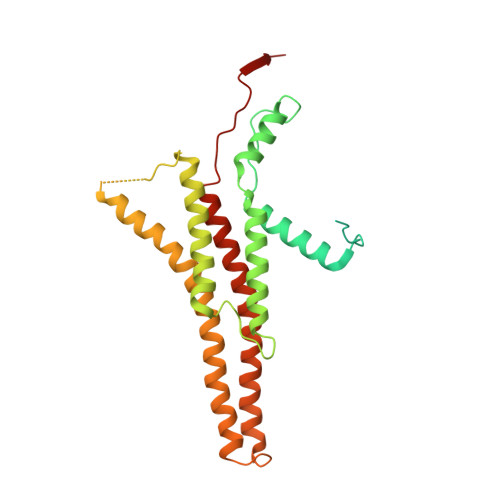
Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Murphy, B.J., Klusch, N., Langer, J., Mills, D.J., Yildiz, O., Kuhlbrandt, W.(2019) Science 364
- PubMed: 31221832
- DOI: https://doi.org/10.1126/science.aaw9128
- Primary Citation of Related Structures:
6RD4, 6RD5, 6RD6, 6RD7, 6RD8, 6RD9, 6RDA, 6RDB, 6RDC, 6RDD, 6RDE, 6RDF, 6RDG, 6RDH, 6RDI, 6RDJ, 6RDK, 6RDL, 6RDM, 6RDN, 6RDO, 6RDP, 6RDQ, 6RDR, 6RDS, 6RDT, 6RDU, 6RDV, 6RDW, 6RDX, 6RDY, 6RDZ, 6RE0, 6RE1, 6RE2, 6RE3, 6RE4, 6RE5, 6RE6, 6RE7, 6RE8, 6RE9, 6REA, 6REB, 6REC, 6RED, 6REE, 6REF, 6REP, 6RER - PubMed Abstract:
F 1 F o -adenosine triphosphate (ATP) synthases make the energy of the proton-motive force available for energy-consuming processes in the cell. We determined the single-particle cryo-electron microscopy structure of active dimeric ATP synthase from mitochondria of Polytomella sp. at a resolution of 2.7 to 2.8 angstroms. Separation of 13 well-defined rotary substates by three-dimensional classification provides a detailed picture of the molecular motions that accompany c -ring rotation and result in ATP synthesis. Crucially, the F 1 head rotates along with the central stalk and c -ring rotor for the first ~30° of each 120° primary rotary step to facilitate flexible coupling of the stoichiometrically mismatched F 1 and F o subcomplexes. Flexibility is mediated primarily by the interdomain hinge of the conserved OSCP subunit. A conserved metal ion in the proton access channel may synchronize c -ring protonation with rotation.
- Department of Structural Biology, Max Planck Institute of Biophysics, Frankfurt 60438, Germany.
Organizational Affiliation: