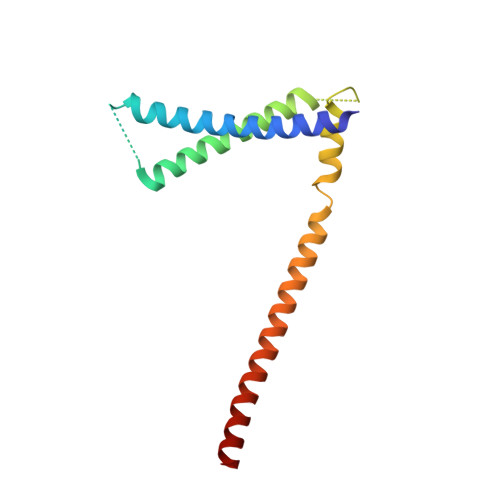
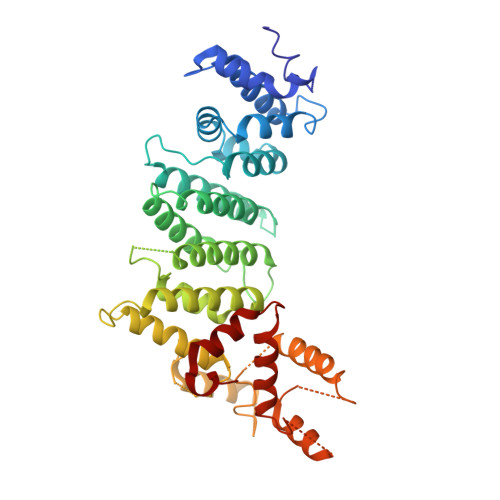
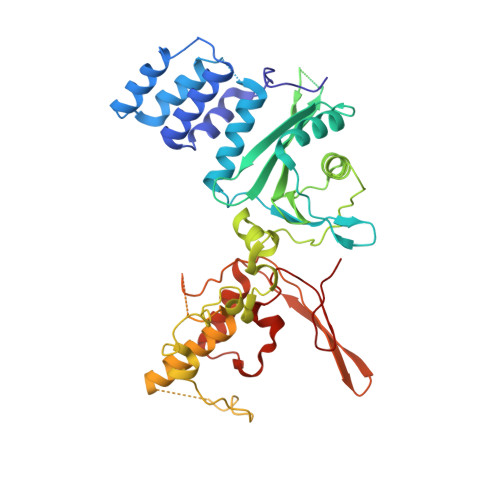
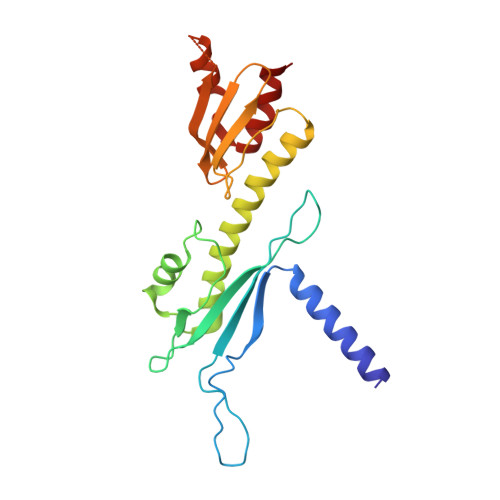
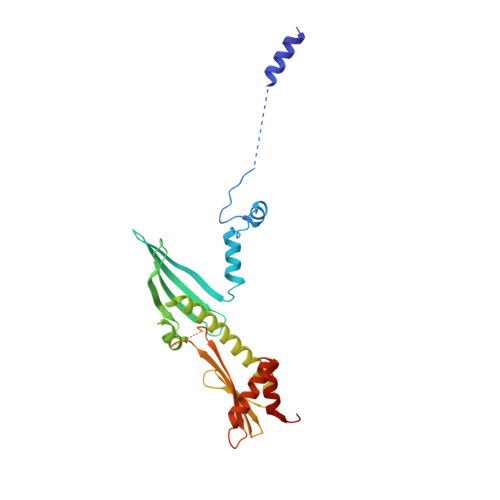
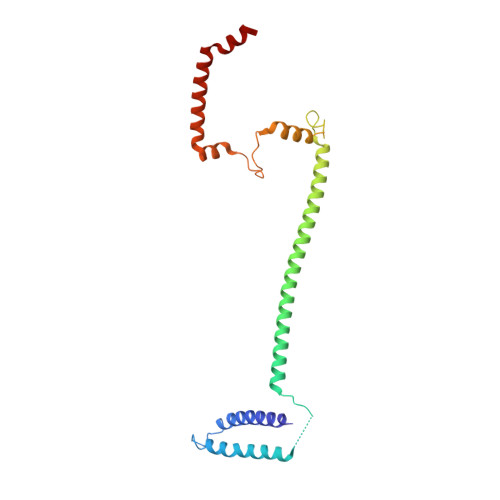
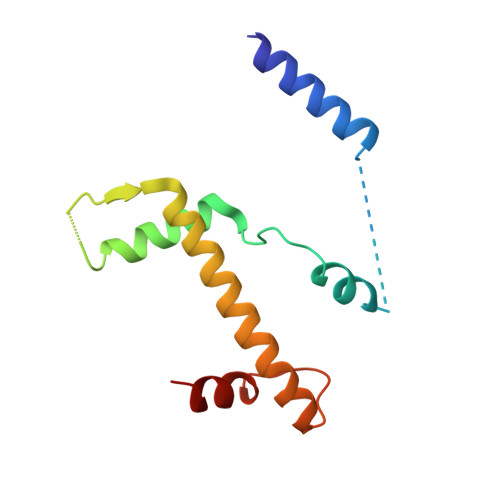
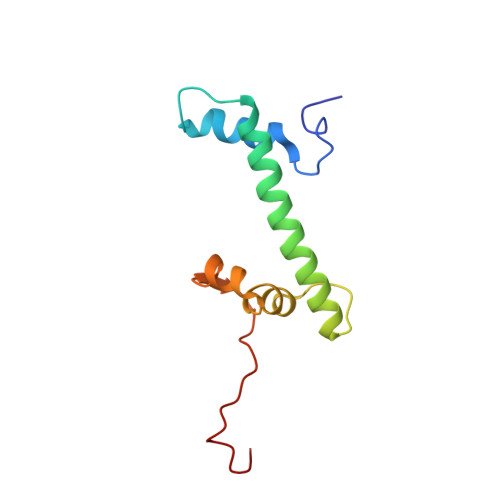
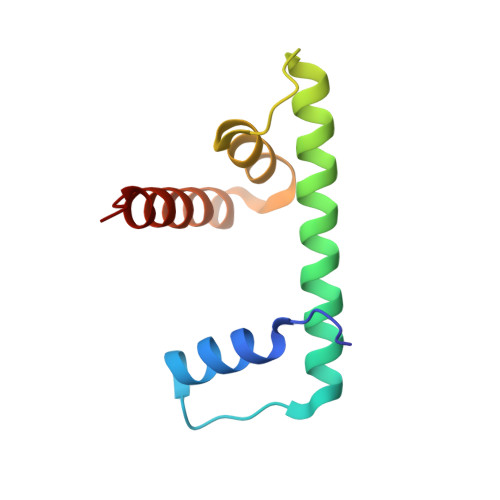
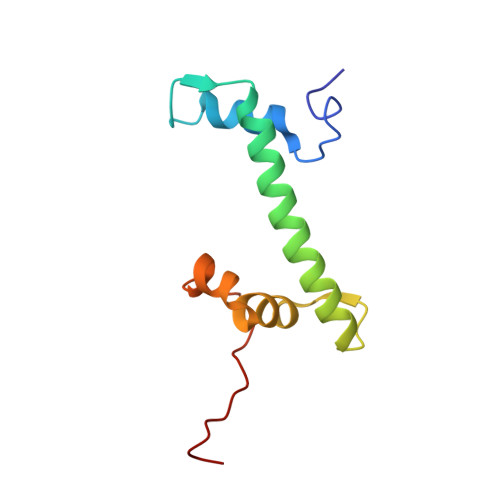
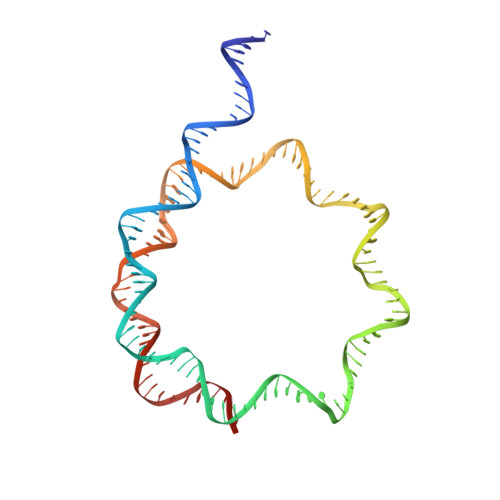
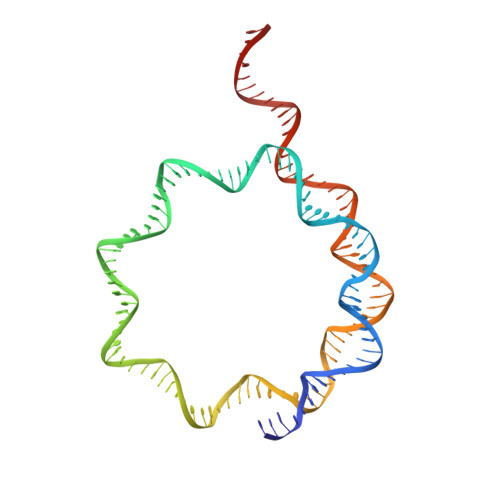
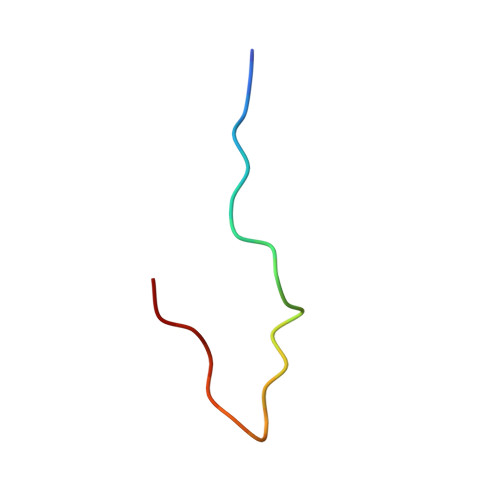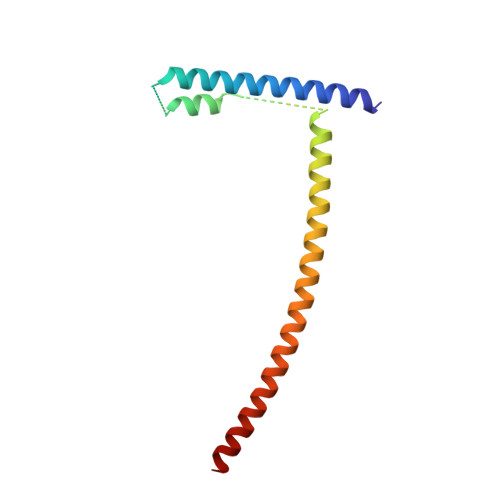Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Yan, K., Yang, J., Zhang, Z., McLaughlin, S.H., Chang, L., Fasci, D., Ehrenhofer-Murray, A.E., Heck, A.J.R., Barford, D.(2019) Nature 574: 278-282
- PubMed: 31578520
- DOI: https://doi.org/10.1038/s41586-019-1609-1
- Primary Citation of Related Structures:
6QLD, 6QLE, 6QLF - PubMed Abstract:
In eukaryotes, accurate chromosome segregation in mitosis and meiosis maintains genome stability and prevents aneuploidy. Kinetochores are large protein complexes that, by assembling onto specialized Cenp-A nucleosomes 1,2 , function to connect centromeric chromatin to microtubules of the mitotic spindle 3,4 . Whereas the centromeres of vertebrate chromosomes comprise millions of DNA base pairs and attach to multiple microtubules, the simple point centromeres of budding yeast are connected to individual microtubules 5,6 . All 16 budding yeast chromosomes assemble complete kinetochores using a single Cenp-A nucleosome (Cenp-A Nuc ), each of which is perfectly centred on its cognate centromere 7-9 . The inner and outer kinetochore modules are responsible for interacting with centromeric chromatin and microtubules, respectively. Here we describe the cryo-electron microscopy structure of the Saccharomyces cerevisiae inner kinetochore module, the constitutive centromere associated network (CCAN) complex, assembled onto a Cenp-A nucleosome (CCAN-Cenp-A Nuc ). The structure explains the interdependency of the constituent subcomplexes of CCAN and shows how the Y-shaped opening of CCAN accommodates Cenp-A Nuc to enable specific CCAN subunits to contact the nucleosomal DNA and histone subunits. Interactions with the unwrapped DNA duplex at the two termini of Cenp-A Nuc are mediated predominantly by a DNA-binding groove in the Cenp-L-Cenp-N subcomplex. Disruption of these interactions impairs assembly of CCAN onto Cenp-A Nuc . Our data indicate a mechanism of Cenp-A nucleosome recognition by CCAN and how CCAN acts as a platform for assembly of the outer kinetochore to link centromeres to the mitotic spindle for chromosome segregation.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: