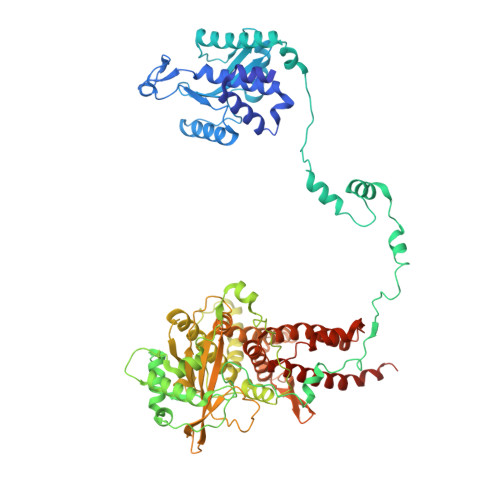
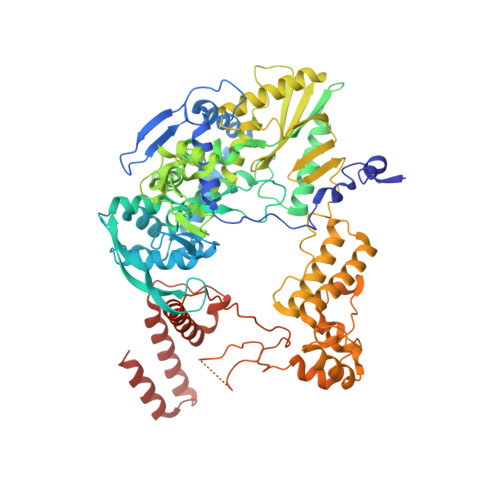
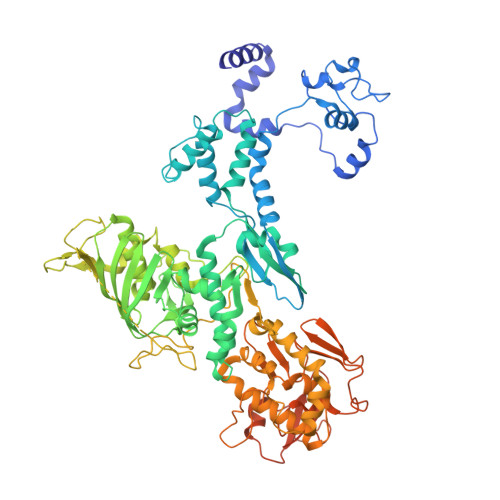
Structural snapshots of actively transcribing influenza polymerase.
Kouba, T., Drncova, P., Cusack, S.(2019) Nat Struct Mol Biol 26: 460-470
- PubMed: 31160782
- DOI: https://doi.org/10.1038/s41594-019-0232-z
- Primary Citation of Related Structures:
6QCS, 6QCT, 6QCV, 6QCW, 6QCX - PubMed Abstract:
Influenza virus RNA-dependent RNA polymerase uses unique mechanisms to transcribe its single-stranded genomic viral RNA (vRNA) into messenger RNA. The polymerase is initially bound to a promoter comprising the partially base-paired 3' and 5' extremities of the RNA. A short, capped primer, 'cap-snatched' from a nascent host polymerase II transcript, is directed towards the polymerase active site to initiate RNA synthesis. Here we present structural snapshots, as determined by X-ray crystallography and cryo-electron microscopy, of actively initiating influenza polymerase as it transitions towards processive elongation. Unexpected conformational changes unblock the active site cavity to allow establishment of a nine-base-pair template-product RNA duplex before the strands separate into distinct exit channels. Concomitantly, as the template translocates, the promoter base pairs are broken and the template entry region is remodeled. These structures reveal details of the influenza polymerase active site that will help optimize nucleoside analogs or other compounds that directly inhibit viral RNA synthesis.
- European Molecular Biology Laboratory, Grenoble, France.
Organizational Affiliation: