Assessment of the nucleotide modifications in the high-resolution cryo-electron microscopy structure of the Escherichia coli 50S subunit.
Stojkovic, V., Myasnikov, A.G., Young, I.D., Frost, A., Fraser, J.S., Fujimori, D.G.(2020) Nucleic Acids Res 48: 2723-2732
- PubMed: 31989172
- DOI: https://doi.org/10.1093/nar/gkaa037
- Primary Citation of Related Structures:
6PJ6 - PubMed Abstract:
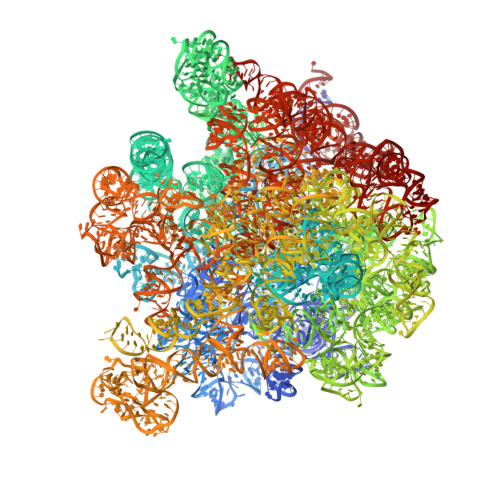
Post-transcriptional ribosomal RNA (rRNA) modifications are present in all organisms, but their exact functional roles and positions are yet to be fully characterized. Modified nucleotides have been implicated in the stabilization of RNA structure and regulation of ribosome biogenesis and protein synthesis. In some instances, rRNA modifications can confer antibiotic resistance. High-resolution ribosome structures are thus necessary for precise determination of modified nucleotides' positions, a task that has previously been accomplished by X-ray crystallography. Here, we present a cryo-electron microscopy (cryo-EM) structure of the Escherichia coli 50S subunit at an average resolution of 2.2 Å as an additional approach for mapping modification sites. Our structure confirms known modifications present in 23S rRNA and additionally allows for localization of Mg2+ ions and their coordinated water molecules. Using our cryo-EM structure as a testbed, we developed a program for assessment of cryo-EM map quality. This program can be easily used on any RNA-containing cryo-EM structure, and an associated Coot plugin allows for visualization of validated modifications, making it highly accessible.
- Department of Cellular and Molecular Pharmacology, University of California San Francisco, San Francisco, CA 94158, USA.
Organizational Affiliation: