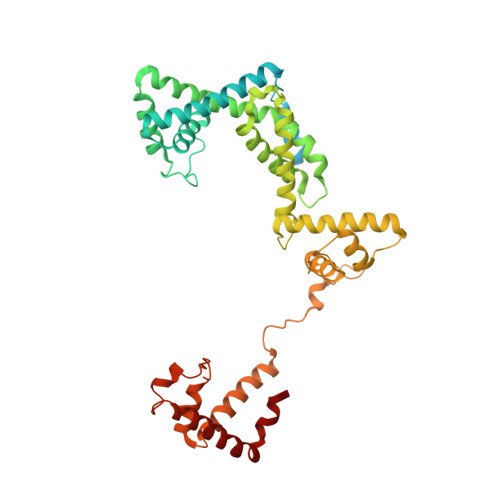
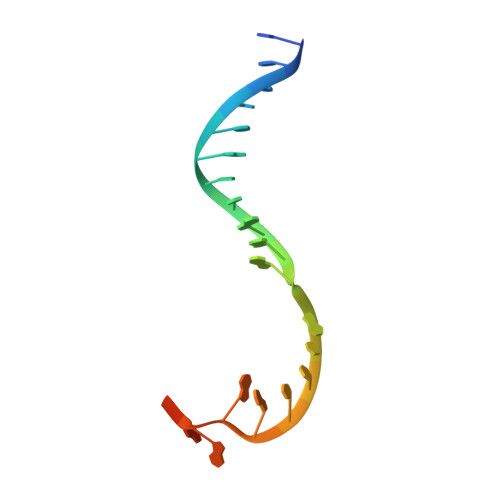
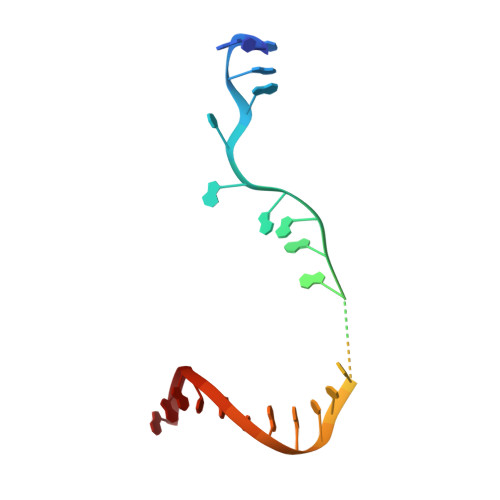
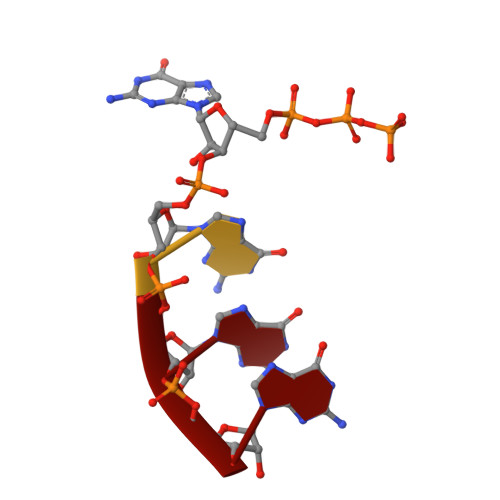
Structural basis of reiterative transcription from the pyrG and pyrBI promoters by bacterial RNA polymerase.
Shin, Y., Hedglin, M., Murakami, K.S.(2020) Nucleic Acids Res 48: 2144-2155
- PubMed: 31965171
- DOI: https://doi.org/10.1093/nar/gkz1221
- Primary Citation of Related Structures:
6OVR, 6OVY, 6OW3, 6OY5, 6OY6, 6OY7, 6P70, 6P71 - PubMed Abstract:
Reiterative transcription is a non-canonical form of RNA synthesis by RNA polymerase in which a ribonucleotide specified by a single base in the DNA template is repetitively added to the nascent RNA transcript. We previously determined the X-ray crystal structure of the bacterial RNA polymerase engaged in reiterative transcription from the pyrG promoter, which contains eight poly-G RNA bases synthesized using three C bases in the DNA as a template and extends RNA without displacement of the promoter recognition σ factor from the core enzyme. In this study, we determined a series of transcript initiation complex structures from the pyrG promoter using soak-trigger-freeze X-ray crystallography. We also performed biochemical assays to monitor template DNA translocation during RNA synthesis from the pyrG promoter and in vitro transcription assays to determine the length of poly-G RNA from the pyrG promoter variants. Our study revealed how RNA slips on template DNA and how RNA polymerase and template DNA determine length of reiterative RNA product. Lastly, we determined a structure of a transcript initiation complex at the pyrBI promoter and proposed an alternative mechanism of RNA slippage and extension requiring the σ dissociation from the core enzyme.
- Department of Biochemistry and Molecular Biology, Center for RNA Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA.
Organizational Affiliation: