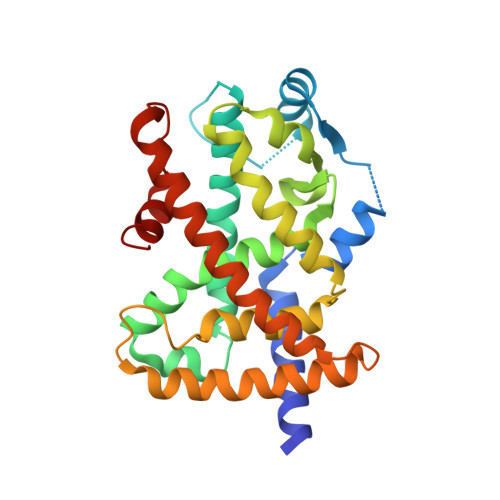
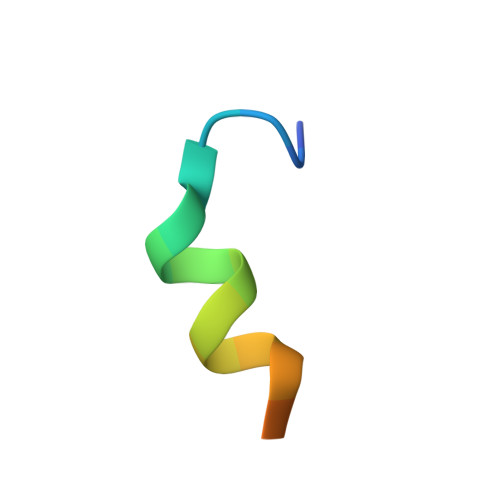A molecular switch regulating transcriptional repression and activation of PPAR gamma.
Shang, J., Mosure, S.A., Zheng, J., Brust, R., Bass, J., Nichols, A., Solt, L.A., Griffin, P.R., Kojetin, D.J.(2020) Nat Commun 11: 956-956
- PubMed: 32075969
- DOI: https://doi.org/10.1038/s41467-020-14750-x
- Primary Citation of Related Structures:
6ONI, 6ONJ, 6PDZ - PubMed Abstract:
Nuclear receptor (NR) transcription factors use a conserved activation function-2 (AF-2) helix 12 mechanism for agonist-induced coactivator interaction and NR transcriptional activation. In contrast, ligand-induced corepressor-dependent NR repression appears to occur through structurally diverse mechanisms. We report two crystal structures of peroxisome proliferator-activated receptor gamma (PPARγ) in an inverse agonist/corepressor-bound transcriptionally repressive conformation. Helix 12 is displaced from the solvent-exposed active conformation and occupies the orthosteric ligand-binding pocket enabled by a conformational change that doubles the pocket volume. Paramagnetic relaxation enhancement (PRE) NMR and chemical crosslinking mass spectrometry confirm the repressive helix 12 conformation. PRE NMR also defines the mechanism of action of the corepressor-selective inverse agonist T0070907, and reveals that apo-helix 12 exchanges between transcriptionally active and repressive conformations-supporting a fundamental hypothesis in the NR field that helix 12 exchanges between transcriptionally active and repressive conformations.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, Jupiter, FL, 33458, USA.
Organizational Affiliation: