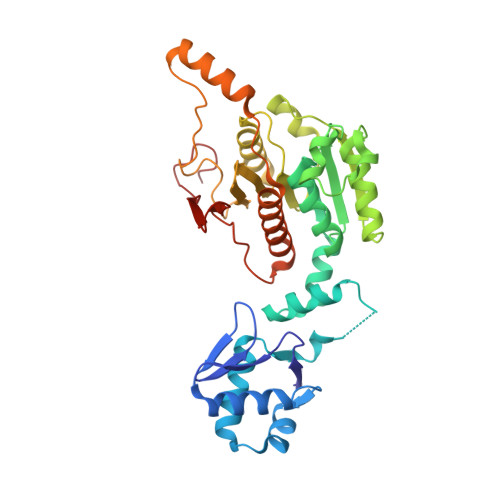
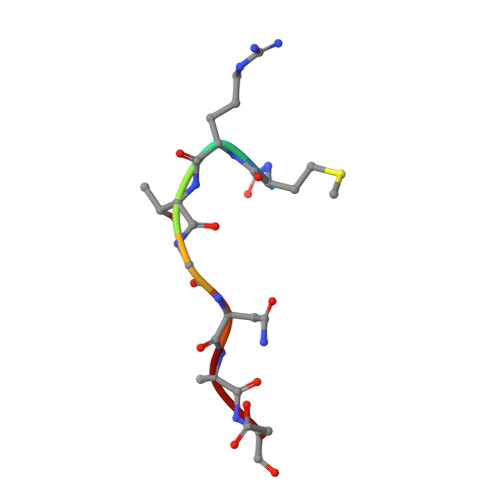
Biosynthesis of the RiPP trojan horse nucleotide antibiotic microcin C is directed by theN-formyl of the peptide precursor.
Dong, S.H., Kulikovsky, A., Zukher, I., Estrada, P., Dubiley, S., Severinov, K., Nair, S.K.(2019) Chem Sci 10: 2391-2395
- PubMed: 30881667
- DOI: https://doi.org/10.1039/c8sc03173h
- Primary Citation of Related Structures:
6OM4 - PubMed Abstract:
Microcin C7 (McC) is a peptide antibiotic modified by a linkage of the terminal isoAsn amide to AMP via a phosphoramidate bond. Post-translational modification on this ribosomally produced heptapeptide precursor is carried out by MccB, which consumes two equivalents of ATP to generate the N-P linkage. We demonstrate that MccB only efficiently processes the precursor heptapeptide that retains the N -formylated initiator Met (fMet). Binding studies and kinetic measurements evidence the role of the N -formyl moiety. Structural data show that the N -formyl peptide binding results in an ordering of residues in the MccB "crossover loop", which dictates specificity in homologous ubiquitin activating enzymes. The N -formyl peptide exhibits substrate inhibition, and cannot be displaced from MccB by the desformyl counterpart. Such substrate inhibition may be a strategy to avert unwanted McC buildup and avert toxicity in the cytoplasm of producing organisms.
- Department of Biochemistry , University of Illinois at Urbana-Champaign , Illinois , USA . Email: snair@illinois.edu.
Organizational Affiliation: