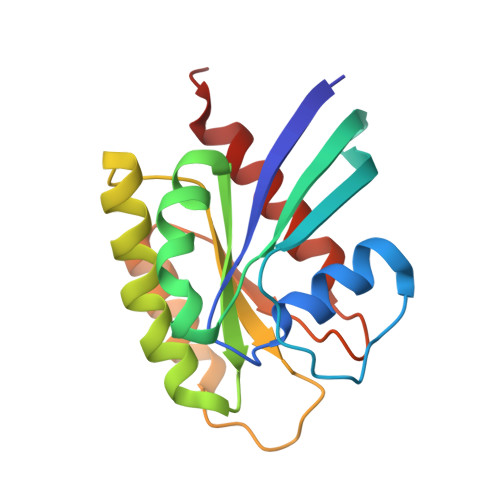
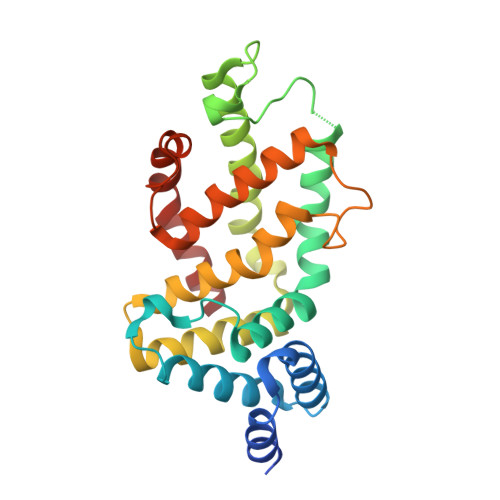
KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Rabara, D., Tran, T.H., Dharmaiah, S., Stephens, R.M., McCormick, F., Simanshu, D.K., Holderfield, M.(2019) Proc Natl Acad Sci U S A 116: 22122-22131
- PubMed: 31611389
- DOI: https://doi.org/10.1073/pnas.1908353116
- Primary Citation of Related Structures:
6OB2, 6OB3 - PubMed Abstract:
KRAS mutations occur in ∼35% of colorectal cancers and promote tumor growth by constitutively activating the mitogen-activated protein kinase (MAPK) pathway. KRAS mutations at codons 12, 13, or 61 are thought to prevent GAP protein-stimulated GTP hydrolysis and render KRAS -mutated colorectal cancers unresponsive to epidermal growth factor receptor (EGFR) inhibitors. We report here that KRAS G13-mutated cancer cells are frequently comutated with NF1 GAP but NF1 is rarely mutated in cancers with KRAS codon 12 or 61 mutations. Neurofibromin protein (encoded by the NF1 gene) hydrolyzes GTP directly in complex with KRAS G13D, and KRAS G13D-mutated cells can respond to EGFR inhibitors in a neurofibromin-dependent manner. Structures of the wild type and G13D mutant of KRAS in complex with neurofibromin (RasGAP domain) provide the structural basis for neurofibromin-mediated GTP hydrolysis. These results reveal that KRAS G13D is responsive to neurofibromin-stimulated hydrolysis and suggest that a subset of KRAS G13-mutated colorectal cancers that are neurofibromin-competent may respond to EGFR therapies.
- NCI RAS Initiative, Cancer Research Technology Program, Frederick National Laboratory for Cancer Research, Frederick, MD 21701.
Organizational Affiliation: