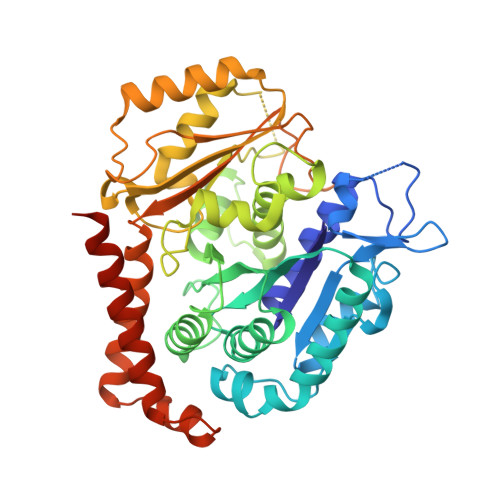
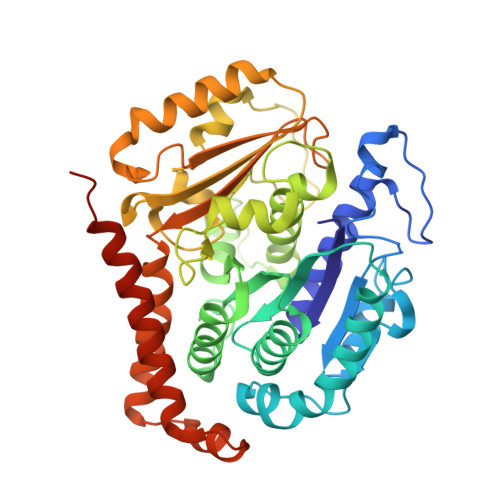
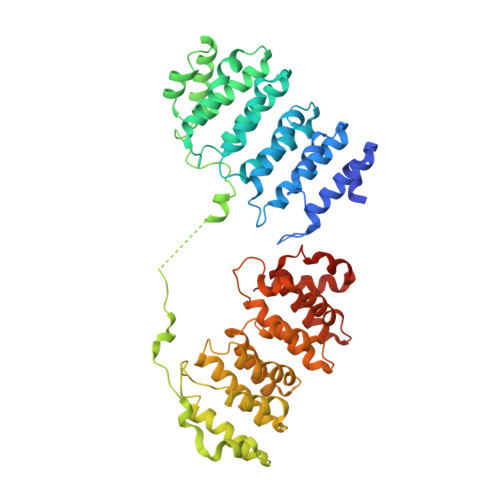
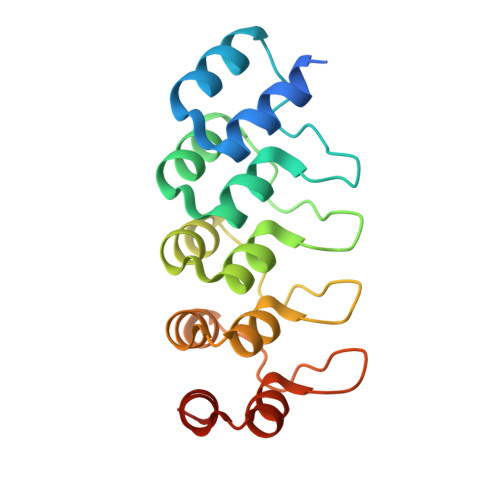
Structural basis of tubulin recruitment and assembly by microtubule polymerases with Tumor Overexpressed Gene (TOG) domain arrays.
Nithianantham, S., Cook, B.D., Beans, M., Guo, F., Chang, F., Al-Bassam, J.(2018) Elife 7
- PubMed: 30422110
- DOI: https://doi.org/10.7554/eLife.38922
- Primary Citation of Related Structures:
6MZE, 6MZF, 6MZG - PubMed Abstract:
XMAP215/Stu2/Alp14 proteins accelerate microtubule plus-end polymerization by recruiting tubulins via arrays of tumor overexpressed gene (TOG) domains, yet their mechanism remains unknown. Here, we describe the biochemical and structural basis for TOG arrays in recruiting and polymerizing tubulins. Alp14 binds four tubulins via dimeric TOG1-TOG2 subunits, in which each domain exhibits a distinct exchange rate for tubulin. X-ray structures revealed square-shaped assemblies composed of pseudo-dimeric TOG1-TOG2 subunits assembled head-to-tail, positioning four unpolymerized tubulins in a polarized wheel-like configuration. Crosslinking and electron microscopy show Alp14-tubulin forms square assemblies in solution, and inactivating their interfaces destabilize this organization without influencing tubulin binding. An X-ray structure determined using approach to modulate tubulin polymerization revealed an unfurled assembly, in which TOG1-TOG2 uniquely bind to two polymerized tubulins. Our findings suggest a new microtubule polymerase model in which TOG arrays recruit tubulins by forming square assemblies that then unfurl, facilitating their concerted polymerization into protofilaments.
- Molecular Cellular Biology Department, University of California, Davis, United States.
Organizational Affiliation: