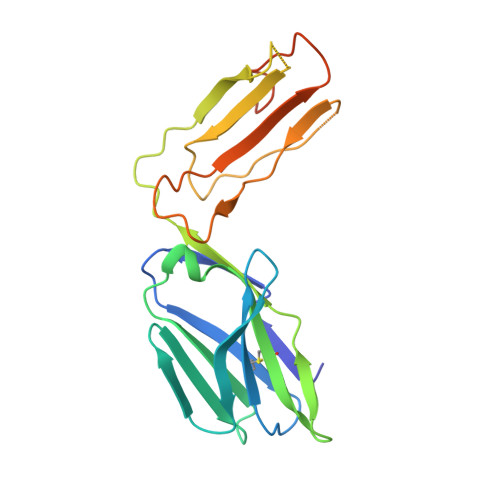
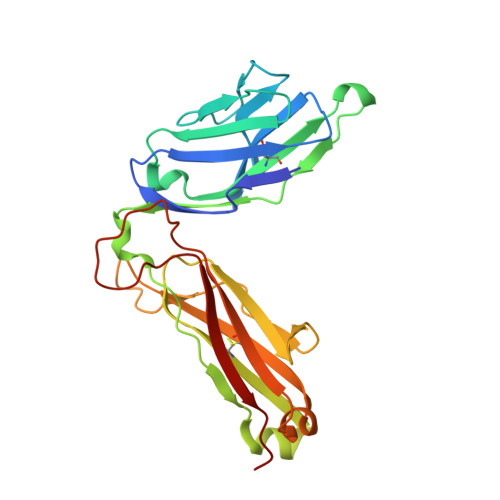
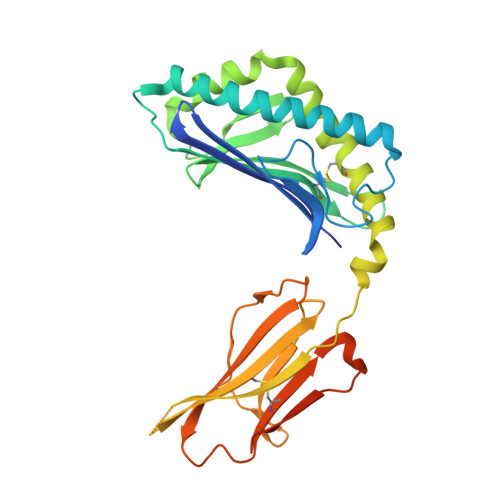
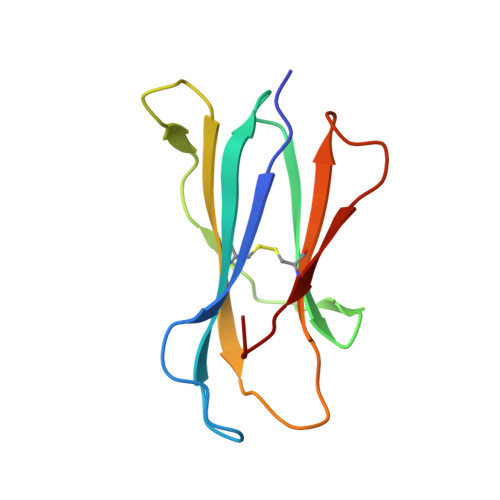
Distinct CD1d docking strategies exhibited by diverse Type II NKT cell receptors.
Almeida, C.F., Sundararaj, S., Le Nours, J., Praveena, T., Cao, B., Burugupalli, S., Smith, D.G.M., Patel, O., Brigl, M., Pellicci, D.G., Williams, S.J., Uldrich, A.P., Godfrey, D.I., Rossjohn, J.(2019) Nat Commun 10: 5242-5242
- PubMed: 31748533
- DOI: https://doi.org/10.1038/s41467-019-12941-9
- Primary Citation of Related Structures:
6MRA, 6MSS - PubMed Abstract:
Type I and type II natural killer T (NKT) cells are restricted to the lipid antigen-presenting molecule CD1d. While we have an understanding of the antigen reactivity and function of type I NKT cells, our knowledge of type II NKT cells in health and disease remains unclear. Here we describe a population of type II NKT cells that recognise and respond to the microbial antigen, α-glucuronosyl-diacylglycerol (α-GlcADAG) presented by CD1d, but not the prototypical type I NKT cell agonist, α-galactosylceramide. Surprisingly, the crystal structure of a type II NKT TCR-CD1d-α-GlcADAG complex reveals a CD1d F'-pocket-docking mode that contrasts sharply with the previously determined A'-roof positioning of a sulfatide-reactive type II NKT TCR. Our data also suggest that diverse type II NKT TCRs directed against distinct microbial or mammalian lipid antigens adopt multiple recognition strategies on CD1d, thereby maximising the potential for type II NKT cells to detect different lipid antigens.
- Department of Microbiology & Immunology, Peter Doherty Institute for Infection and Immunity, University of Melbourne, Melbourne, VIC, 3010, Australia.
Organizational Affiliation: