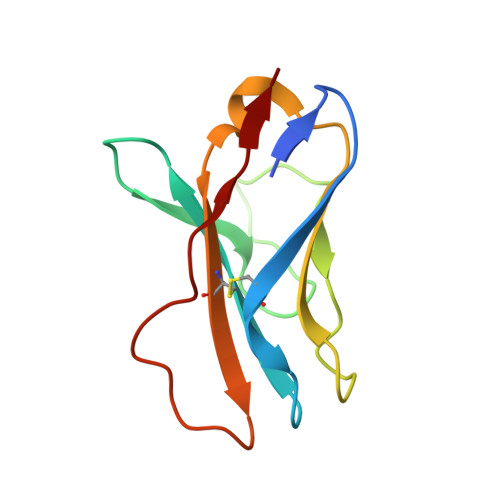
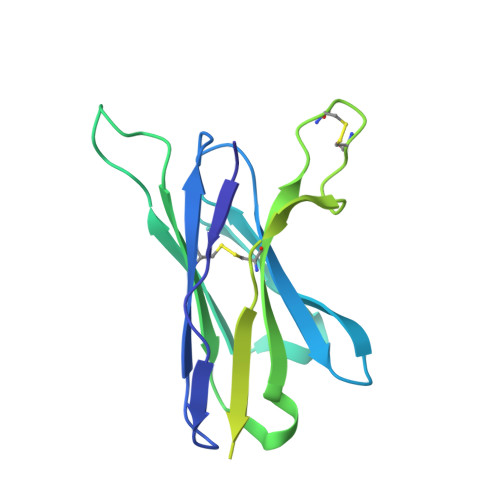
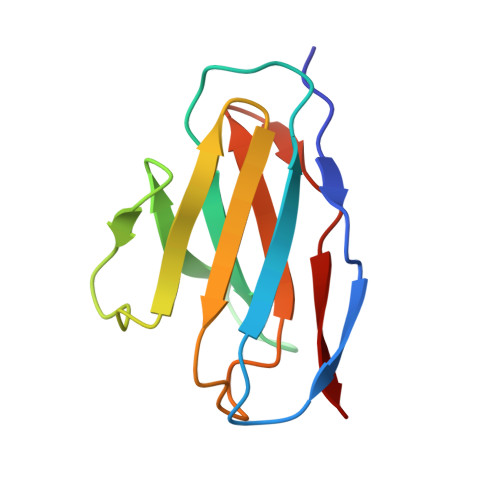
Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Kong, R., Duan, H., Sheng, Z., Xu, K., Acharya, P., Chen, X., Cheng, C., Dingens, A.S., Gorman, J., Sastry, M., Shen, C.H., Zhang, B., Zhou, T., Chuang, G.Y., Chao, C.W., Gu, Y., Jafari, A.J., Louder, M.K., O'Dell, S., Rowshan, A.P., Viox, E.G., Wang, Y., Choi, C.W., Corcoran, M.M., Corrigan, A.R., Dandey, V.P., Eng, E.T., Geng, H., Foulds, K.E., Guo, Y., Kwon, Y.D., Lin, B., Liu, K., Mason, R.D., Nason, M.C., Ohr, T.Y., Ou, L., Rawi, R., Sarfo, E.K., Schon, A., Todd, J.P., Wang, S., Wei, H., Wu, W., Mullikin, J.C., Bailer, R.T., Doria-Rose, N.A., Karlsson Hedestam, G.B., Scorpio, D.G., Overbaugh, J., Bloom, J.D., Carragher, B., Potter, C.S., Shapiro, L., Kwong, P.D., Mascola, J.R.(2019) Cell 178: 567-584.e19
- PubMed: 31348886
- DOI: https://doi.org/10.1016/j.cell.2019.06.030
- Primary Citation of Related Structures:
6MPG, 6MPH, 6MQC, 6MQE, 6MQM, 6MQR, 6MQS, 6N16, 6N1V, 6N1W, 6NF2, 6OSY, 6OT1 - PubMed Abstract:
The vaccine-mediated elicitation of antibodies (Abs) capable of neutralizing diverse HIV-1 strains has been a long-standing goal. To understand how broadly neutralizing antibodies (bNAbs) can be elicited, we identified, characterized, and tracked five neutralizing Ab lineages targeting the HIV-1-fusion peptide (FP) in vaccinated macaques over time. Genetic and structural analyses revealed two of these lineages to belong to a reproducible class capable of neutralizing up to 59% of 208 diverse viral strains. B cell analysis indicated each of the five lineages to have been initiated and expanded by FP-carrier priming, with envelope (Env)-trimer boosts inducing cross-reactive neutralization. These Abs had binding-energy hotspots focused on FP, whereas several FP-directed Abs induced by immunization with Env trimer-only were less FP-focused and less broadly neutralizing. Priming with a conserved subregion, such as FP, can thus induce Abs with binding-energy hotspots coincident with the target subregion and capable of broad neutralization.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, MD 20892, USA.
Organizational Affiliation: