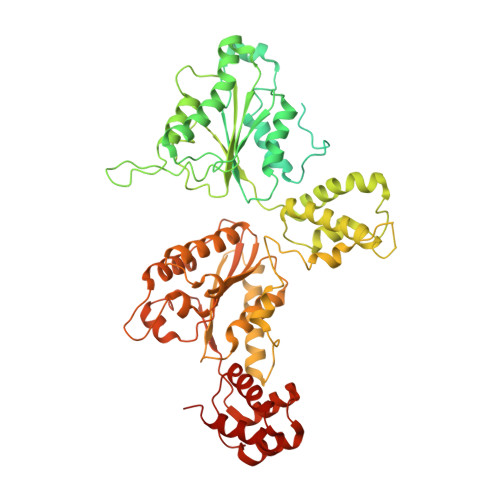
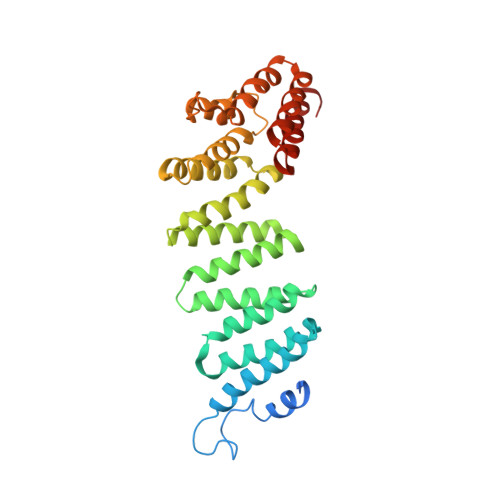
Structural principles of SNARE complex recognition by the AAA+ protein NSF.
White, K.I., Zhao, M., Choi, U.B., Pfuetzner, R.A., Brunger, A.T.(2018) Elife 7
- PubMed: 30198481
- DOI: https://doi.org/10.7554/eLife.38888
- Primary Citation of Related Structures:
6MDM, 6MDN, 6MDO, 6MDP - PubMed Abstract:
The recycling of SNARE proteins following complex formation and membrane fusion is an essential process in eukaryotic trafficking. A highly conserved AAA+ protein, NSF ( N -ethylmaleimide sensitive factor) and an adaptor protein, SNAP (soluble NSF attachment protein), disassemble the SNARE complex. We report electron-cryomicroscopy structures of the complex of NSF, αSNAP, and the full-length soluble neuronal SNARE complex (composed of syntaxin-1A, synaptobrevin-2, SNAP-25A) in the presence of ATP under non-hydrolyzing conditions at ~3.9 Å resolution. These structures reveal electrostatic interactions by which two αSNAP molecules interface with a specific surface of the SNARE complex. This interaction positions the SNAREs such that the 15 N-terminal residues of SNAP-25A are loaded into the D1 ring pore of NSF via a spiral pattern of interactions between a conserved tyrosine NSF residue and SNAP-25A backbone atoms. This loading process likely precedes ATP hydrolysis. Subsequent ATP hydrolysis then drives complete disassembly.
- Department of Molecular and Cellular Physiology, Stanford University, Stanford, United States.
Organizational Affiliation: