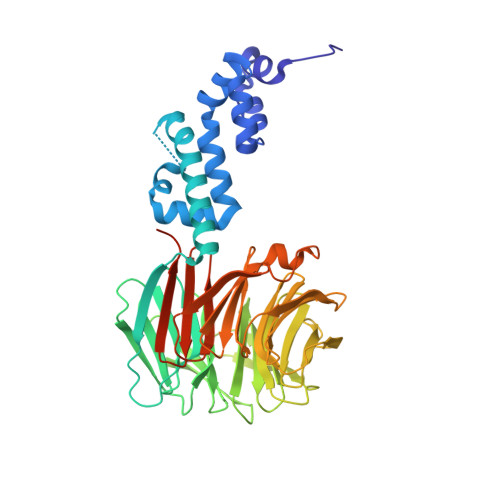
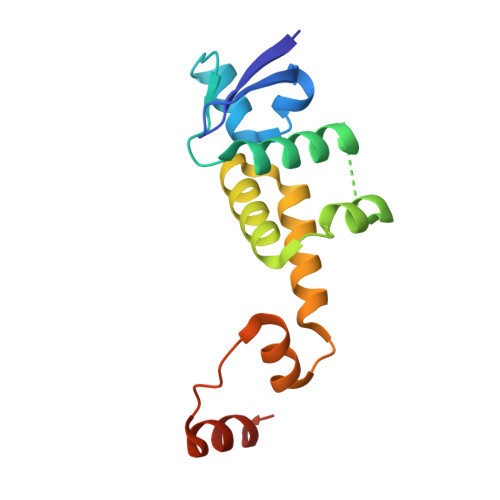
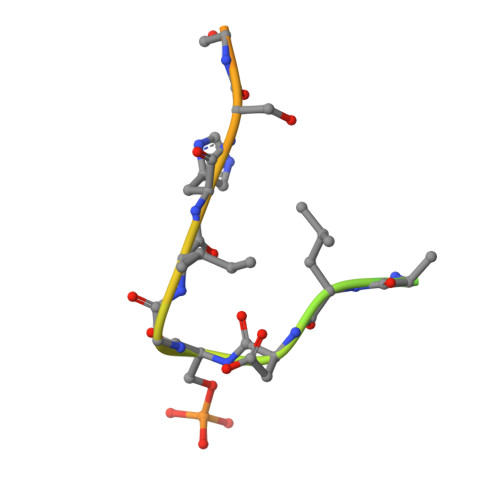
Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction.
Simonetta, K.R., Taygerly, J., Boyle, K., Basham, S.E., Padovani, C., Lou, Y., Cummins, T.J., Yung, S.L., von Soly, S.K., Kayser, F., Kuriyan, J., Rape, M., Cardozo, M., Gallop, M.A., Bence, N.F., Barsanti, P.A., Saha, A.(2019) Nat Commun 10: 1402-1402
- PubMed: 30926793
- DOI: https://doi.org/10.1038/s41467-019-09358-9
- Primary Citation of Related Structures:
6M90, 6M91, 6M92, 6M93, 6M94 - PubMed Abstract:
Protein-protein interactions (PPIs) governing the recognition of substrates by E3 ubiquitin ligases are critical to cellular function. There is significant therapeutic potential in the development of small molecules that modulate these interactions; however, rational design of small molecule enhancers of PPIs remains elusive. Herein, we report the prospective identification and rational design of potent small molecules that enhance the interaction between an oncogenic transcription factor, β-Catenin, and its cognate E3 ligase, SCF β-TrCP . These enhancers potentiate the ubiquitylation of mutant β-Catenin by β-TrCP in vitro and induce the degradation of an engineered mutant β-Catenin in a cellular system. Distinct from PROTACs, these drug-like small molecules insert into a naturally occurring PPI interface, with contacts optimized for both the substrate and ligase within the same small molecule entity. The prospective discovery of 'molecular glue' presented here provides a paradigm for the development of small molecule degraders targeting hard-to-drug proteins.
- Nurix Therapeutics, Inc., 1700 Owens Street, Suite 205, San Francisco, CA, 94158, USA. ksimonetta@nurix-inc.com.
Organizational Affiliation: