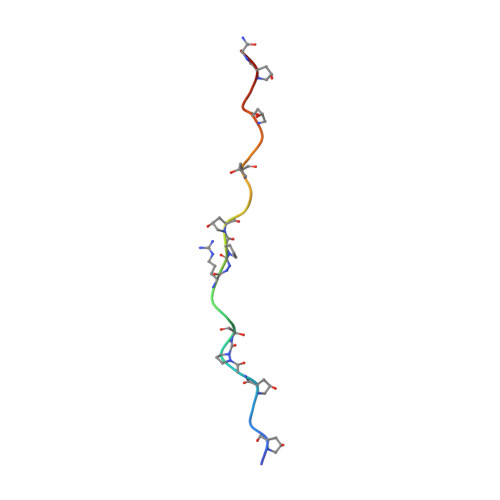
Aza-proline effectively mimics l-proline stereochemistry in triple helical collagen.
Kasznel, A.J., Harris, T., Porter, N.J., Zhang, Y., Chenoweth, D.M.(2019) Chem Sci 10: 6979-6983
- PubMed: 31588264
- DOI: https://doi.org/10.1039/c9sc02211b
- Primary Citation of Related Structures:
6M80 - PubMed Abstract:
The prevalence of l-amino acids in biomolecules has been shown to have teleological importance in biomolecular structure and self-assembly. Recently, biophysical studies have demonstrated that natural l-amino acids can be replaced with non-natural achiral aza-amino acids in folded protein structures such as triple helical collagen. However, the structural consequences of achiral aza-amino acid incorporation has not been elucidated in the context of any relevant folded biomolecule. Herein, we use X-ray crystallography to provide the first atomic resolution crystal structure of an achiral aza-amino acid residue embedded within a folded protein structure, definitively illustrating that achiral aza-proline has the capacity to effectively mimic the stereochemistry of natural amino acids within the context of triple helical collagen. We further corroborate this finding with density functional theory computational analysis showing that the natural l-amino acid stereochemistry for aza-proline is energetically favored when arranged in the aza-proline-hydroxyproline-glycine motif. In addition to providing fundamental insight into peptide and protein structure, the incorporation of achiral stereochemical mimics such as aza-amino acids could have far reaching impacts in areas ranging from synthetic materials to drug design.
- Department of Chemistry , University of Pennsylvania , 231 S. 34th St. , Philadelphia , PA 19104-6323 , USA . Email: dcheno@sas.upenn.edu.
Organizational Affiliation: