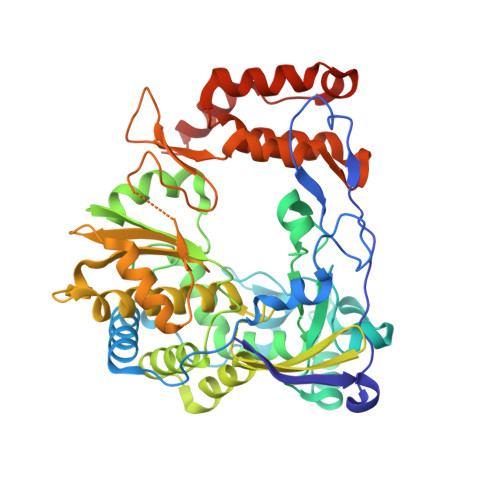
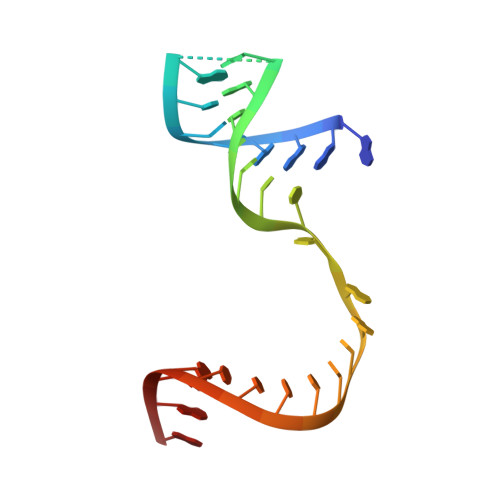
A nucleobase-binding pocket in a viral RNA-dependent RNA polymerase contributes to elongation complex stability.
Shi, W., Ye, H.Q., Deng, C.L., Li, R., Zhang, B., Gong, P.(2020) Nucleic Acids Res 48: 1392-1405
- PubMed: 31863580
- DOI: https://doi.org/10.1093/nar/gkz1170
- Primary Citation of Related Structures:
6KWQ, 6KWR - PubMed Abstract:
The enterovirus 71 (EV71) 3Dpol is an RNA-dependent RNA polymerase (RdRP) that plays the central role in the viral genome replication, and is an important target in antiviral studies. Here, we report a crystal structure of EV71 3Dpol elongation complex (EC) at 1.8 Å resolution. The structure reveals that the 5'-end guanosine of the downstream RNA template interacts with a fingers domain pocket, with the base sandwiched by H44 and R277 side chains through hydrophobic stacking interactions, and these interactions are still maintained after one in-crystal translocation event induced by nucleotide incorporation, implying that the pocket could regulate the functional properties of the polymerase by interacting with RNA. When mutated, residue R277 showed an impact on virus proliferation in virological studies with residue H44 having a synergistic effect. In vitro biochemical data further suggest that mutations at these two sites affect RNA binding, EC stability, but not polymerase catalytic rate (kcat) and apparent NTP affinity (KM,NTP). We propose that, although rarely captured by crystallography, similar surface pocket interaction with nucleobase may commonly exist in nucleic acid motor enzymes to facilitate their processivity. Potential applications in antiviral drug and vaccine development are also discussed.
- Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, No.44 Xiao Hong Shan, Wuhan, Hubei 430071, China.
Organizational Affiliation: