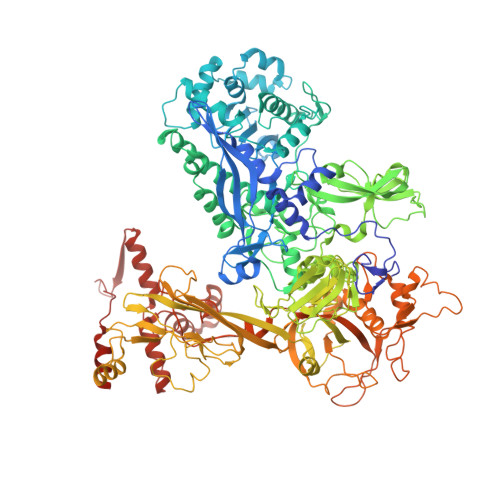
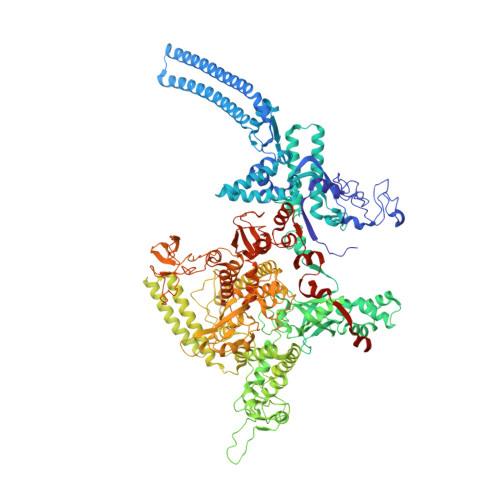
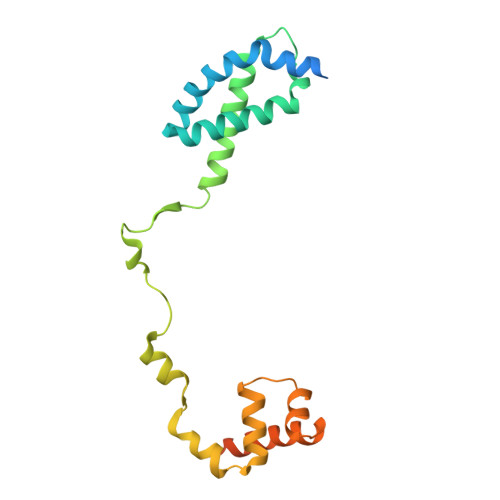
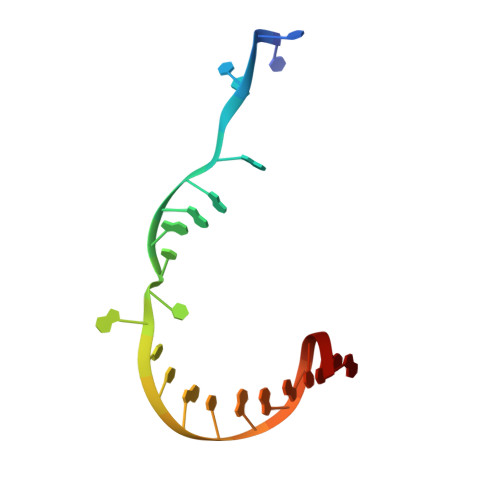
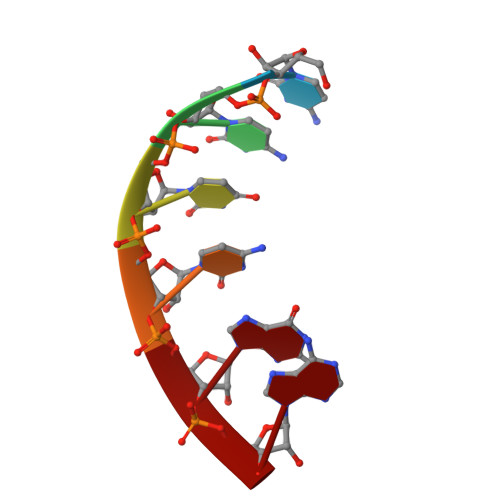
Structures and mechanism of transcription initiation by bacterial ECF factors.
Fang, C., Li, L., Shen, L., Shi, J., Wang, S., Feng, Y., Zhang, Y.(2019) Nucleic Acids Res 47: 7094-7104
- PubMed: 31131408
- DOI: https://doi.org/10.1093/nar/gkz470
- Primary Citation of Related Structures:
6JBQ, 6JCX, 6JCY - PubMed Abstract:
Bacterial RNA polymerase (RNAP) forms distinct holoenzymes with extra-cytoplasmic function (ECF) σ factors to initiate specific gene expression programs. In this study, we report a cryo-EM structure at 4.0 Å of Escherichia coli transcription initiation complex comprising σE-the most-studied bacterial ECF σ factor (Ec σE-RPo), and a crystal structure at 3.1 Å of Mycobacterium tuberculosis transcription initiation complex with a chimeric σH/E (Mtb σH/E-RPo). The structure of Ec σE-RPo reveals key interactions essential for assembly of E. coli σE-RNAP holoenzyme and for promoter recognition and unwinding by E. coli σE. Moreover, both structures show that the non-conserved linkers (σ2/σ4 linker) of the two ECF σ factors are inserted into the active-center cleft and exit through the RNA-exit channel. We performed secondary-structure prediction of 27,670 ECF σ factors and find that their non-conserved linkers probably reach into and exit from RNAP active-center cleft in a similar manner. Further biochemical results suggest that such σ2/σ4 linker plays an important role in RPo formation, abortive production and promoter escape during ECF σ factors-mediated transcription initiation.
- Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Organizational Affiliation: