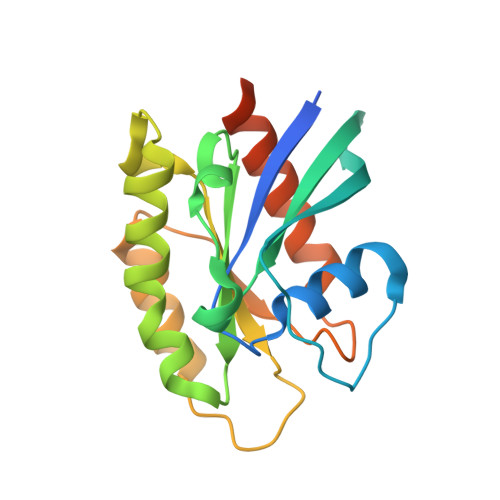
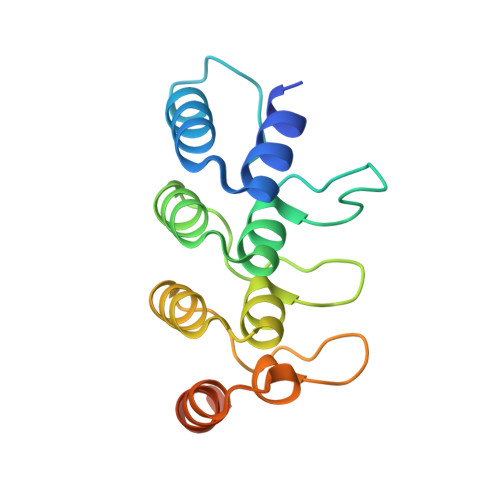
Structural basis of human ORP1-Rab7 interaction for the late-endosome and lysosome targeting.
Tong, J., Tan, L., Chun, C., Im, Y.J.(2019) PLoS One 14: e0211724-e0211724
- PubMed: 30721249
- DOI: https://doi.org/10.1371/journal.pone.0211724
- Primary Citation of Related Structures:
6IYB - PubMed Abstract:
Oxysterol-binding protein (OSBP) and OSBP-related proteins (ORPs) constitute a family of lipid transfer proteins conserved in eukaryotes. ORP1 transports cholesterol at the interface between the late endosomes/lysosomes (LELs) and the endoplasmic reticulum (ER). ORP1 is targeted to the endosomal membranes by forming a tripartite complex with the LE GTPase Rab7 and its effector RILP (Rab7-interacting lysosomal protein). Here, we determined the crystal structure of human ORP1 ANK domain in complex with the GTP-bound form of Rab7. ORP1 ANK binds to the helix α3 of Rab7 located away from the switching regions, which makes the interaction independent of the nucleotide-binding state of Rab7. Thus, the effector-interacting switch regions of Rab7 are accessible for RILP binding, allowing formation of the ORP1-Rab7-RILP complex. ORP1 ANK binds to Rab7 and the Rab7-RILP complex with similar micro-molar affinities, which is consistent with the independence binding of ORP1 and RILP to Rab7. The structural model of the ORP1-Rab7-RILP complex correlates with the recruitment of ORP1 at the LEL-ER interface and the role in lipid transport and regulation.
- College of Pharmacy, Chonnam National University, Gwangju, Republic of Korea.
Organizational Affiliation: