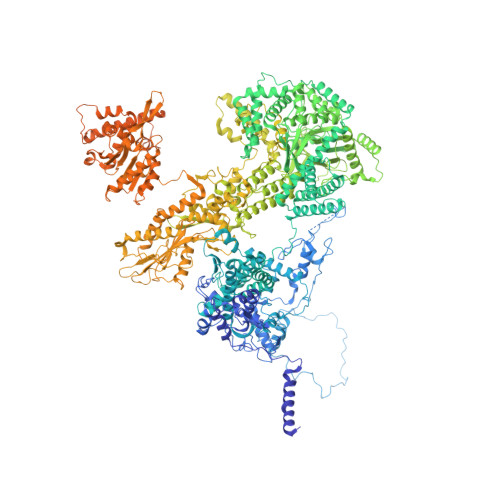
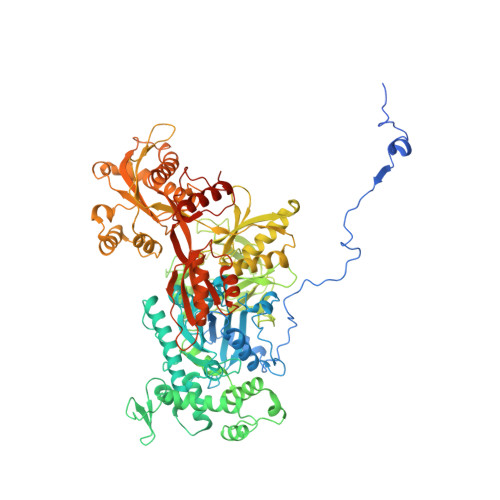
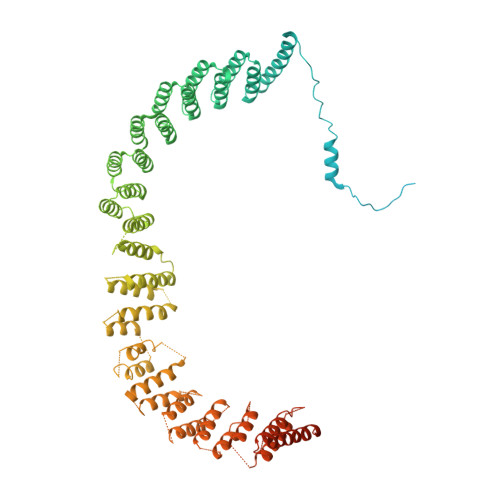
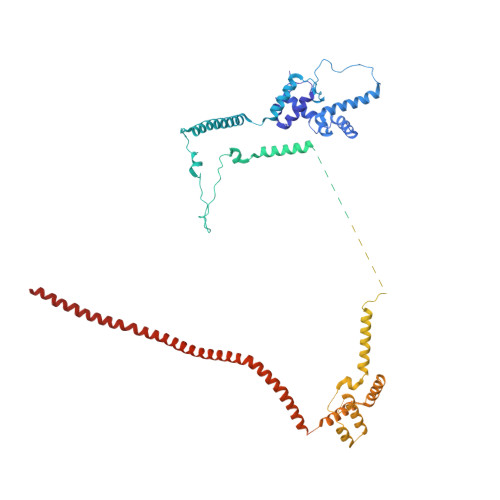
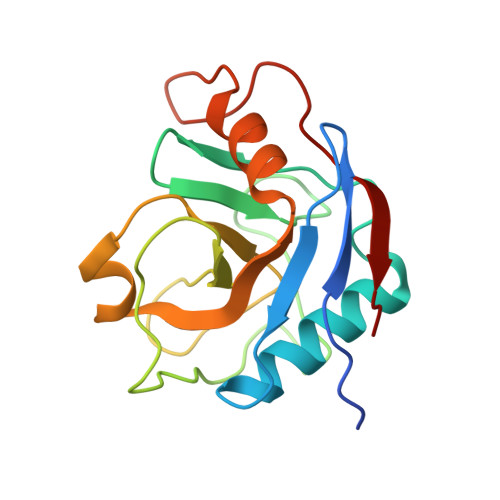
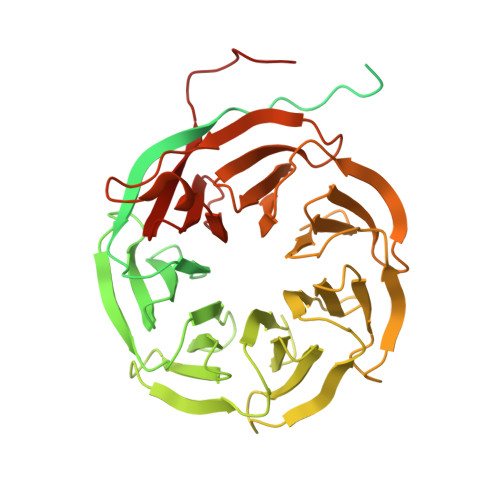
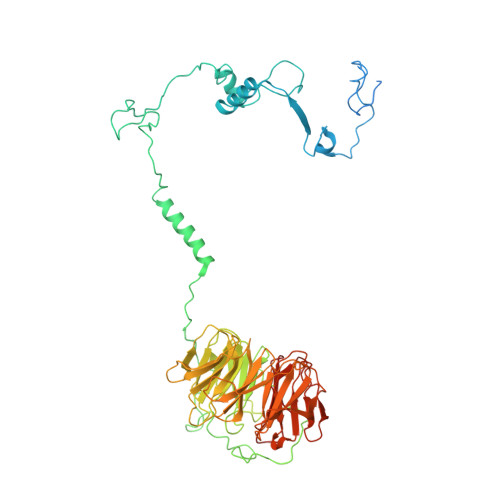
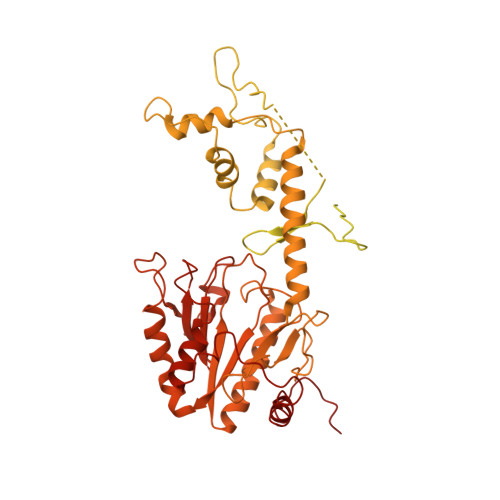
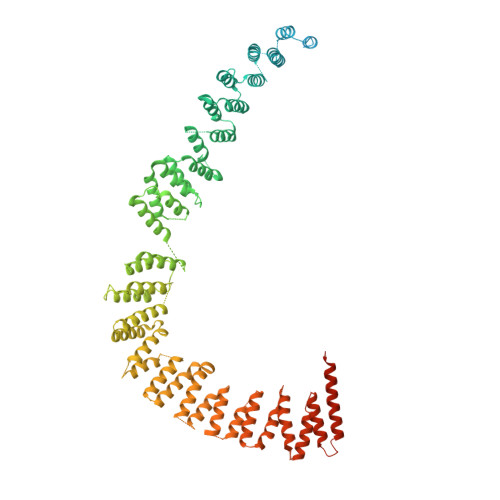
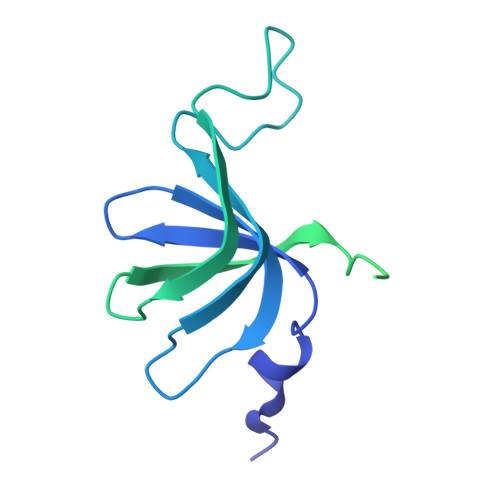
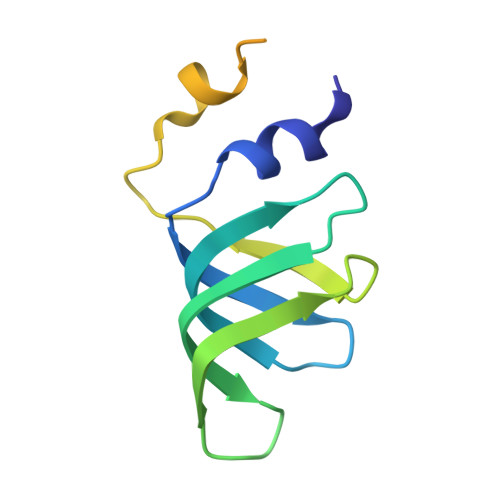
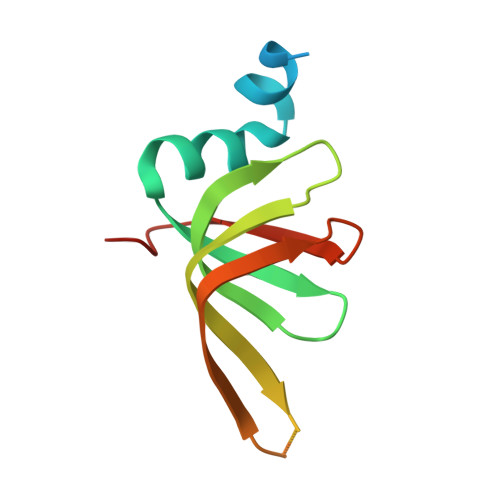
Structures of the human spliceosomes before and after release of the ligated exon.
Zhang, X., Zhan, X., Yan, C., Zhang, W., Liu, D., Lei, J., Shi, Y.(2019) Cell Res 29: 274-285
- PubMed: 30728453
- DOI: https://doi.org/10.1038/s41422-019-0143-x
- Primary Citation of Related Structures:
6ICZ, 6ID0, 6ID1 - PubMed Abstract:
Pre-mRNA splicing is executed by the spliceosome, which has eight major functional states each with distinct composition. Five of these eight human spliceosomal complexes, all preceding exon ligation, have been structurally characterized. In this study, we report the cryo-electron microscopy structures of the human post-catalytic spliceosome (P complex) and intron lariat spliceosome (ILS) at average resolutions of 3.0 and 2.9 Å, respectively. In the P complex, the ligated exon remains anchored to loop I of U5 small nuclear RNA, and the 3'-splice site is recognized by the junction between the 5'-splice site and the branch point sequence. The ATPase/helicase Prp22, along with the ligated exon and eight other proteins, are dissociated in the P-to-ILS transition. Intriguingly, the ILS complex exists in two distinct conformations, one with the ATPase/helicase Prp43 and one without. Comparison of these three late-stage human spliceosomes reveals mechanistic insights into exon release and spliceosome disassembly.
- Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences and School of Medicine, Tsinghua University, Beijing, 100084, China.
Organizational Affiliation: