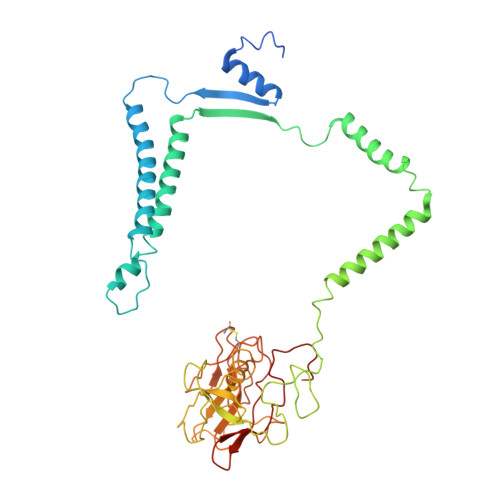
Structure of a functional obligate complex III2IV2respiratory supercomplex from Mycobacterium smegmatis.
Wiseman, B., Nitharwal, R.G., Fedotovskaya, O., Schafer, J., Guo, H., Kuang, Q., Benlekbir, S., Sjostrand, D., Adelroth, P., Rubinstein, J.L., Brzezinski, P., Hogbom, M.(2018) Nat Struct Mol Biol 25: 1128-1136
- PubMed: 30518849
- DOI: https://doi.org/10.1038/s41594-018-0160-3
- Primary Citation of Related Structures:
6HWH - PubMed Abstract:
In the mycobacterial electron-transport chain, respiratory complex III passes electrons from menaquinol to complex IV, which in turn reduces oxygen, the terminal acceptor. Electron transfer is coupled to transmembrane proton translocation, thus establishing the electrochemical proton gradient that drives ATP synthesis. We isolated, biochemically characterized, and determined the structure of the obligate III 2 IV 2 supercomplex from Mycobacterium smegmatis, a model for Mycobacterium tuberculosis. The supercomplex has quinol:O 2 oxidoreductase activity without exogenous cytochrome c and includes a superoxide dismutase subunit that may detoxify reactive oxygen species produced during respiration. We found menaquinone bound in both the Q o and Q i sites of complex III. The complex III-intrinsic diheme cytochrome cc subunit, which functionally replaces both cytochrome c 1 and soluble cytochrome c in canonical electron-transport chains, displays two conformations: one in which it provides a direct electronic link to complex IV and another in which it serves as an electrical switch interrupting the connection.
- Department of Biochemistry and Biophysics, Arrhenius Laboratories for Natural Sciences, Stockholm University, Stockholm, Sweden.
Organizational Affiliation: