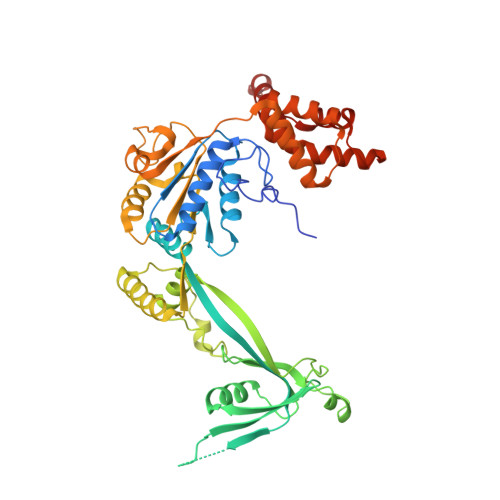
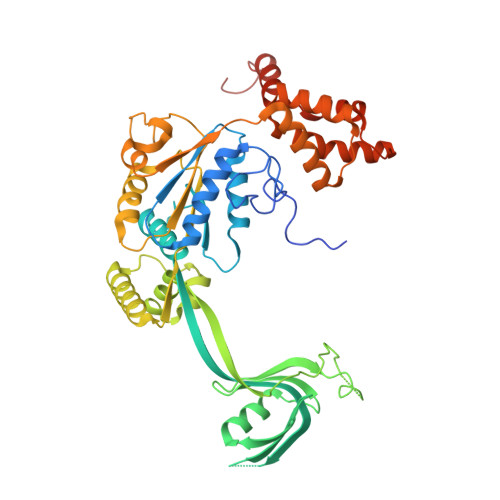
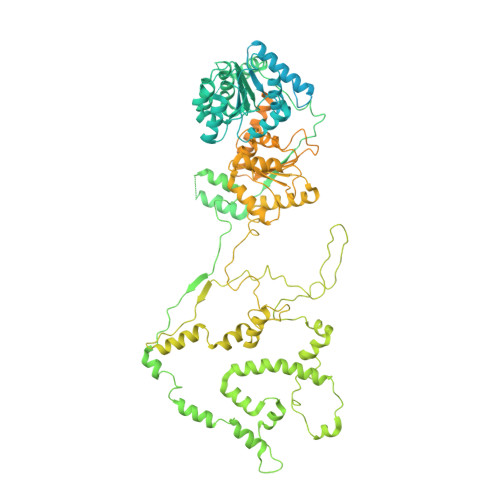
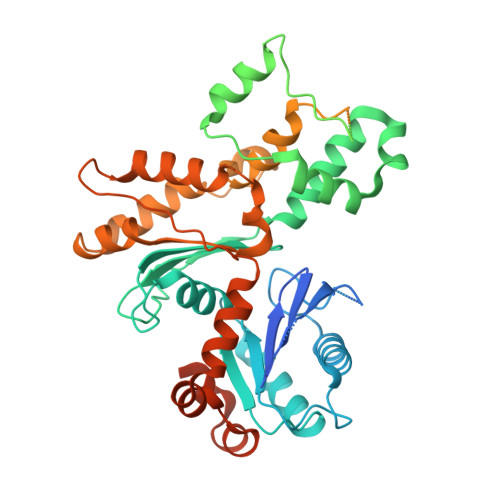
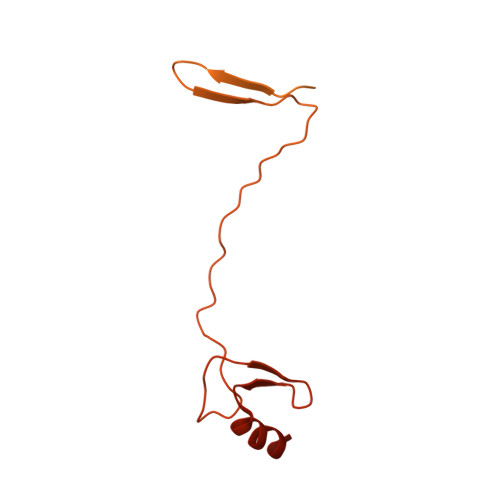
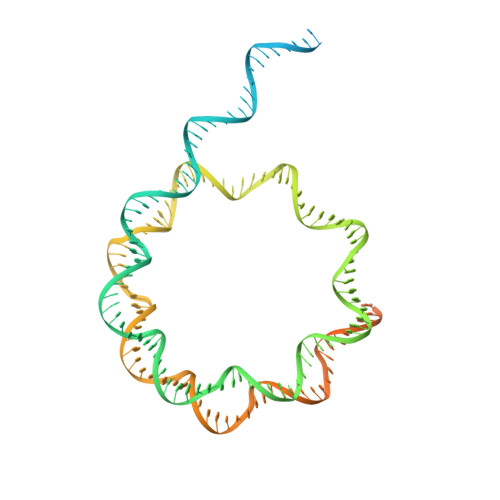
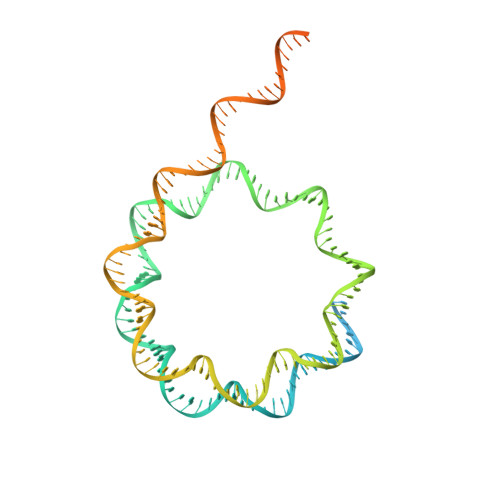
Structure and regulation of the human INO80-nucleosome complex.
Ayala, R., Willhoft, O., Aramayo, R.J., Wilkinson, M., McCormack, E.A., Ocloo, L., Wigley, D.B., Zhang, X.(2018) Nature 556: 391-395
- PubMed: 29643506
- DOI: https://doi.org/10.1038/s41586-018-0021-6
- Primary Citation of Related Structures:
6HTS - PubMed Abstract:
Access to DNA within nucleosomes is required for a variety of processes in cells including transcription, replication and repair. Consequently, cells encode multiple systems that remodel nucleosomes. These complexes can be simple, involving one or a few protein subunits, or more complicated multi-subunit machines 1 . Biochemical studies 2-4 have placed the motor domains of several chromatin remodellers in the superhelical location 2 region of the nucleosome. Structural studies of yeast Chd1 and Snf2-a subunit in the complex with the capacity to remodel the structure of chromatin (RSC)-in complex with nucleosomes 5-7 have provided insights into the basic mechanism of nucleosome sliding performed by these complexes. However, how larger, multi-subunit remodelling complexes such as INO80 interact with nucleosomes and how remodellers carry out functions such as nucleosome sliding 8 , histone exchange 9 and nucleosome spacing 10-12 remain poorly understood. Although some remodellers work as monomers 13 , others work as highly cooperative dimers 11, 14, 15 . Here we present the structure of the human INO80 chromatin remodeller with a bound nucleosome, which reveals that INO80 interacts with nucleosomes in a previously undescribed manner: the motor domains are located on the DNA at the entry point to the nucleosome, rather than at superhelical location 2. The ARP5-IES6 module of INO80 makes additional contacts on the opposite side of the nucleosome. This arrangement enables the histone H3 tails of the nucleosome to have a role in the regulation of the activities of the INO80 motor domain-unlike in other characterized remodellers, for which H4 tails have been shown to regulate the motor domains.
- Section of Structural Biology, Dept. Medicine, Imperial College London, London, UK.
Organizational Affiliation: