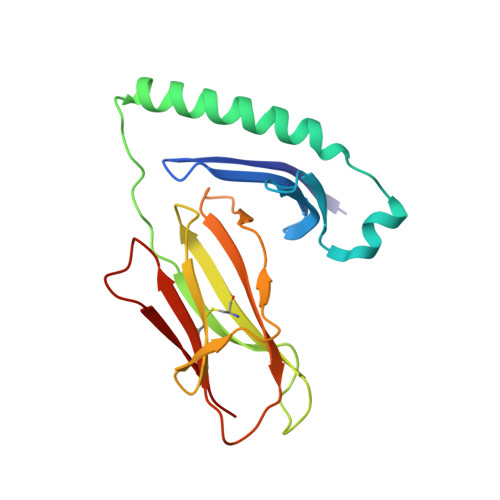
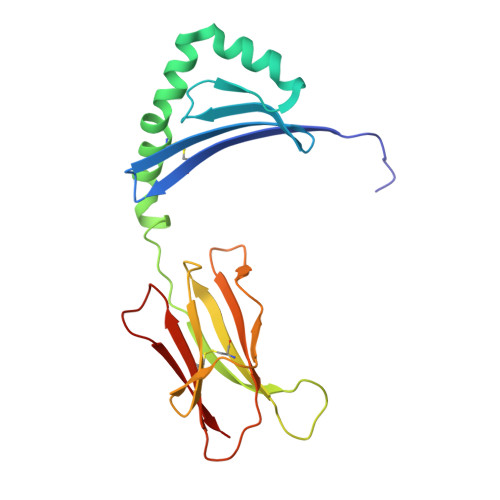
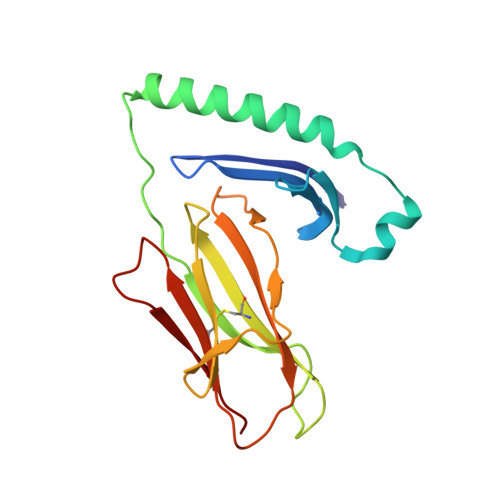
Human leukocyte antigen (HLA) class II peptide flanking residues tune the immunogenicity of a human tumor-derived epitope.
MacLachlan, B.J., Dolton, G., Papakyriakou, A., Greenshields-Watson, A., Mason, G.H., Schauenburg, A., Besneux, M., Szomolay, B., Elliott, T., Sewell, A.K., Gallimore, A., Rizkallah, P., Cole, D.K., Godkin, A.(2019) J Biological Chem 294: 20246-20258
- PubMed: 31619516
- DOI: https://doi.org/10.1074/jbc.RA119.009437
- Primary Citation of Related Structures:
6HBY - PubMed Abstract:
CD4 + T-cells recognize peptide antigens, in the context of human leukocyte antigen (HLA) class II molecules (HLA-II), which through peptide-flanking residues (PFRs) can extend beyond the limits of the HLA binding. The role of the PFRs during antigen recognition is not fully understood; however, recent studies have indicated that these regions can influence T-cell receptor (TCR) affinity and pHLA-II stability. Here, using various biochemical approaches including peptide sensitivity ELISA and ELISpot assays, peptide-binding assays and HLA-II tetramer staining, we focused on CD4 + T-cell responses against a tumor antigen, 5T4 oncofetal trophoblast glycoprotein (5T4), which have been associated with improved control of colorectal cancer. Despite their weak TCR-binding affinity, we found that anti-5T4 CD4 + T-cells are polyfunctional and that their PFRs are essential for TCR recognition of the core bound nonamer. The high-resolution (1.95 Å) crystal structure of HLA-DR1 presenting the immunodominant 20-mer peptide 5T4 111-130 , combined with molecular dynamic simulations, revealed how PFRs explore the HLA-proximal space to contribute to antigen reactivity. These findings advance our understanding of what constitutes an HLA-II epitope and indicate that PFRs can tune weak affinity TCR-pHLA-II interactions.
- Division of Infection and Immunity and Systems Immunity Research Institute, Cardiff University, Cardiff CF14 4XN, United Kingdom.
Organizational Affiliation: