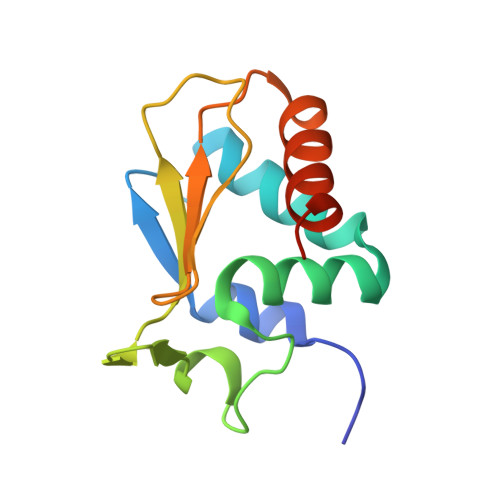
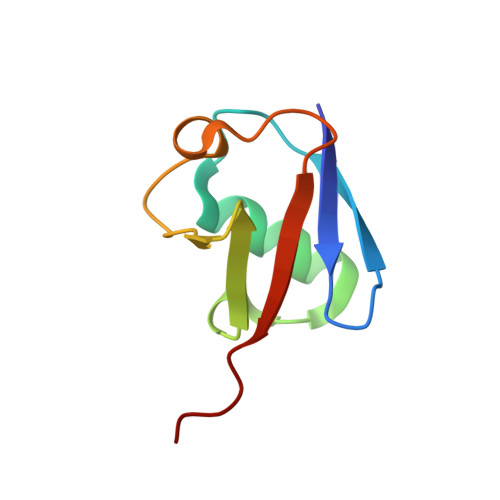
beta-Sheet Augmentation Is a Conserved Mechanism of Priming HECT E3 Ligases for Ubiquitin Ligation.
Jackl, M., Stollmaier, C., Strohaker, T., Hyz, K., Maspero, E., Polo, S., Wiesner, S.(2018) J Mol Biology 430: 3218-3233
- PubMed: 29964046
- DOI: https://doi.org/10.1016/j.jmb.2018.06.044
- Primary Citation of Related Structures:
6FX4, 6FYH - PubMed Abstract:
Ubiquitin (Ub) ligases (E3s) catalyze the attachment of Ub chains to target proteins and thereby regulate a wide array of signal transduction pathways in eukaryotes. In HECT-type E3s, Ub first forms a thioester intermediate with a strictly conserved Cys in the C-lobe of the HECT domain and is then ligated via an isopeptide bond to a Lys residue in the substrate or a preceding Ub in a poly-Ub chain. To date, many key aspects of HECT-mediated Ub transfer have remained elusive. Here, we provide structural and functional insights into the catalytic mechanism of the HECT-type ligase Huwe1 and compare it to the unrelated, K63-specific Smurf2 E3, a member of the Nedd4 family. We found that the Huwe1 HECT domain, in contrast to Nedd4-family E3s, prioritizes K6- and K48-poly-Ub chains and does not interact with Ub in a non-covalent manner. Despite these mechanistic differences, we demonstrate that the architecture of the C-lobe~Ub intermediate is conserved between Huwe1 and Smurf2 and involves a reorientation of the very C-terminal residues. Moreover, in Nedd4 E3s and Huwe1, the individual sequence composition of the Huwe1 C-terminal tail modulates ubiquitination activity, without affecting thioester formation. In sum, our data suggest that catalysis of HECT ligases hold common features, such as the β-sheet augmentation that primes the enzymes for ligation, and variable elements, such as the sequence of the HECT C-terminal tail, that fine-tune ubiquitination activity and may aid in determining Ub chain specificity by positioning the substrate or acceptor Ub.
- Max Planck Institute for Developmental Biology, Max-Planck-Ring 5, 72076 Tübingen, Germany.
Organizational Affiliation: