Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Yang, K., Stanfield, R.L., Martinez-Yamout, M.A., Dyson, H.J., Wilson, I.A., Wright, P.E.(2018) Proc Natl Acad Sci U S A 115: 10040-10045
- PubMed: 30232260
- DOI: https://doi.org/10.1073/pnas.1810397115
- Primary Citation of Related Structures:
6DMX, 6DNQ - PubMed Abstract:
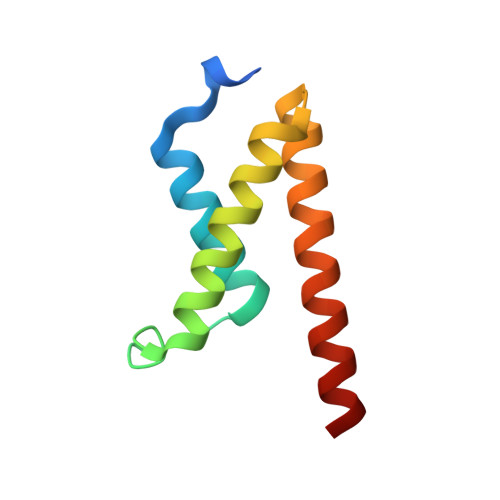
The human T cell leukemia virus I basic leucine zipper protein (HTLV-1 HBZ) maintains chronic viral infection and promotes leukemogenesis through poorly understood mechanisms involving interactions with the KIX domain of the transcriptional coactivator CBP and its paralog p300. The KIX domain binds regulatory proteins at the distinct MLL and c-Myb/pKID sites to form binary or ternary complexes. The intrinsically disordered N-terminal activation domain of HBZ (HBZ AD) deregulates cellular signaling pathways by competing directly with cellular and viral transcription factors for binding to the MLL site and by allosterically perturbing binding of the transactivation domain of the hematopoietic transcription factor c-Myb. Crystal structures of the ternary KIX:c-Myb:HBZ complex show that the HBZ AD recruits two KIX:c-Myb entities through tandem amphipathic motifs (L/V)(V/L)DGLL and folds into a long α-helix upon binding. Isothermal titration calorimetry reveals strong cooperativity in binding of the c-Myb activation domain to the KIX:HBZ complex and in binding of HBZ to the KIX:c-Myb complex. In addition, binding of KIX to the two HBZ (V/L)DGLL motifs is cooperative; the structures suggest that this cooperativity is achieved through propagation of the HBZ α-helix beyond the first binding motif. Our study suggests that the unique structural flexibility and the multiple interaction motifs of the intrinsically disordered HBZ AD are responsible for its potency in hijacking KIX-mediated transcription pathways. The KIX:c-Myb:HBZ complex provides an example of cooperative stabilization in a transcription factor:coactivator network and gives insights into potential mechanisms through which HBZ dysregulates hematopoietic transcriptional programs and promotes T cell proliferation.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037.
Organizational Affiliation: