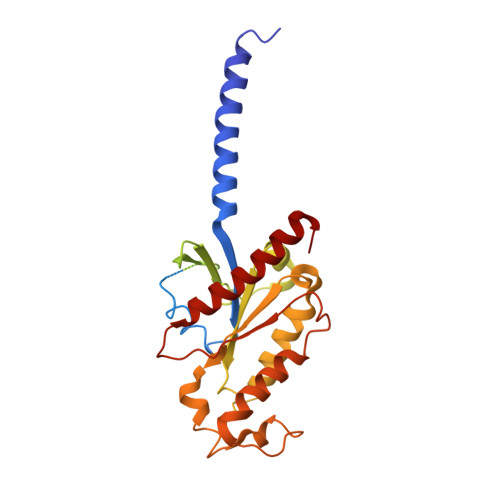
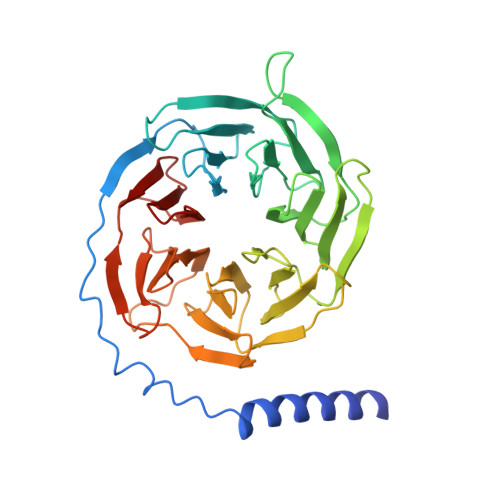
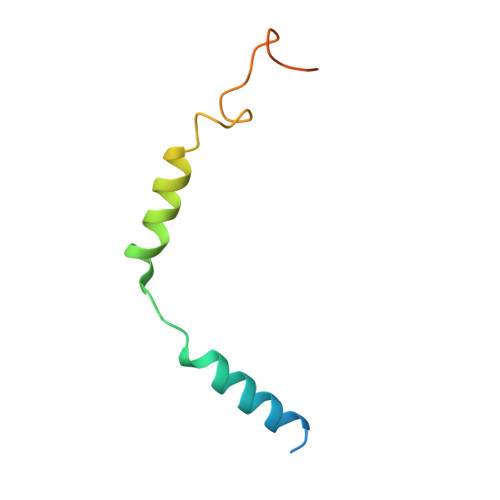
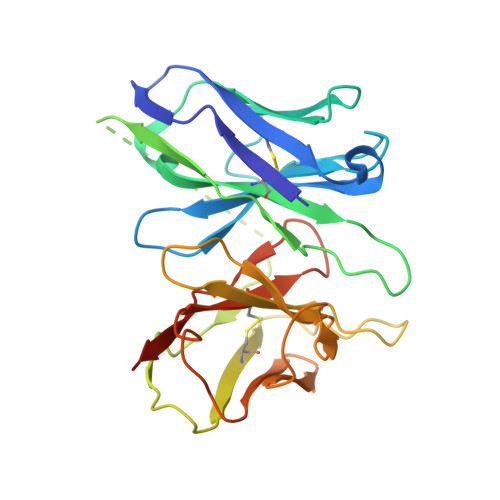
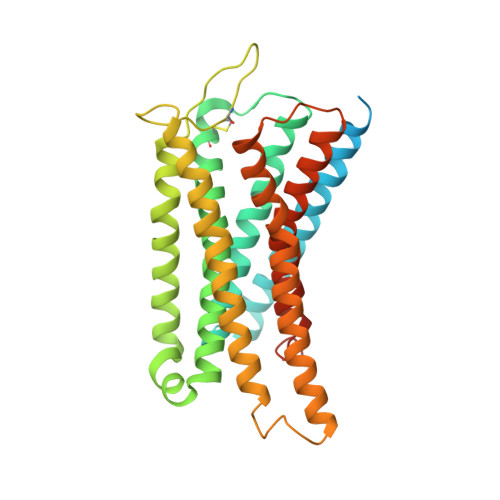
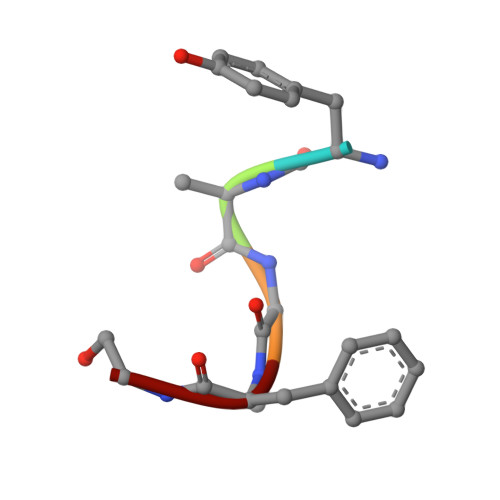
Structure of the mu-opioid receptor-Giprotein complex.
Koehl, A., Hu, H., Maeda, S., Zhang, Y., Qu, Q., Paggi, J.M., Latorraca, N.R., Hilger, D., Dawson, R., Matile, H., Schertler, G.F.X., Granier, S., Weis, W.I., Dror, R.O., Manglik, A., Skiniotis, G., Kobilka, B.K.(2018) Nature 558: 547-552
- PubMed: 29899455
- DOI: https://doi.org/10.1038/s41586-018-0219-7
- Primary Citation of Related Structures:
6DDE, 6DDF - PubMed Abstract:
The μ-opioid receptor (μOR) is a G-protein-coupled receptor (GPCR) and the target of most clinically and recreationally used opioids. The induced positive effects of analgesia and euphoria are mediated by μOR signalling through the adenylyl cyclase-inhibiting heterotrimeric G protein G i . Here we present the 3.5 Å resolution cryo-electron microscopy structure of the μOR bound to the agonist peptide DAMGO and nucleotide-free G i . DAMGO occupies the morphinan ligand pocket, with its N terminus interacting with conserved receptor residues and its C terminus engaging regions important for opioid-ligand selectivity. Comparison of the μOR-G i complex to previously determined structures of other GPCRs bound to the stimulatory G protein G s reveals differences in the position of transmembrane receptor helix 6 and in the interactions between the G protein α-subunit and the receptor core. Together, these results shed light on the structural features that contribute to the G i protein-coupling specificity of the µOR.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, CA, USA.
Organizational Affiliation: