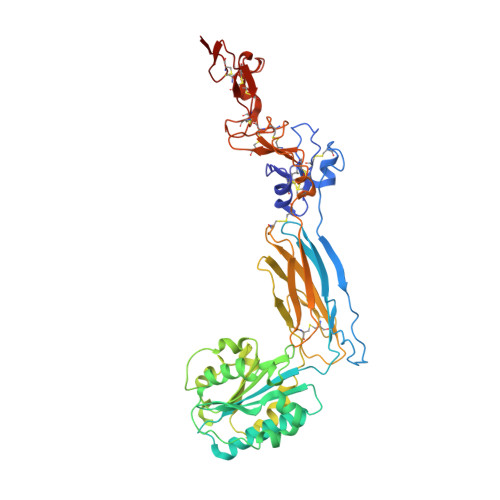
Structure of an extended beta3integrin.
Zhou, D., Thinn, A.M.M., Zhao, Y., Wang, Z., Zhu, J.(2018) Blood 132: 962-972
- PubMed: 30018079
- DOI: https://doi.org/10.1182/blood-2018-01-829572
- Primary Citation of Related Structures:
6BXB, 6BXF, 6CKB - PubMed Abstract:
Cells use adhesion receptor integrins to communicate with their surroundings. Integrin activation and cellular signaling are coupled with change from bent to extended conformation. β 3 integrins, including α IIb β 3 , which is essential for the function of platelets in hemostasis and thrombosis, and α V β 3 , which plays multiple roles in diverse cell types, have been prototypes in understanding integrin structure and function. Despite extensive structural studies, a high-resolution integrin structure in an extended conformation remains to be determined. The human β 3 Leu33Pro polymorphism, located at the PSI domain, defines human platelet-specific alloantigens 1a and 1b (HPA-1a/b), immune response to which is a cause of posttransfusion purpura and fetal/neonatal alloimmune thrombocytopenia. Leu33Pro substitution has also been suggested to be a risk factor for thrombosis. Here we report the crystal structure of the β 3 headpiece in either Leu33 or Pro33 form, both of which reveal intermediate and fully extended conformations coexisting in 1 crystal. These were used to build high-resolution structures of full-length β 3 integrin in the intermediate and fully extended states, agreeing well with the corresponding conformations observed by electron microscopy. Our structures reveal how β 3 integrin becomes extended at its β-knee region and how the flexibility of β-leg domains is determined. In addition, our structures reveal conformational changes of the PSI and I-EGF1 domains upon β 3 extension, which may affect the binding of conformation-dependent anti-HPA-1a alloantibodies. Our structural and functional data show that Leu33Pro substitution does not directly alter the conformation or ligand binding of β 3 integrin.
- Blood Research Institute, BloodCenter of Wisconsin, part of Versiti, Milwaukee, WI.
Organizational Affiliation: