HIV-1 vaccine design through minimizing envelope metastability.
He, L., Kumar, S., Allen, J.D., Huang, D., Lin, X., Mann, C.J., Saye-Francisco, K.L., Copps, J., Sarkar, A., Blizard, G.S., Ozorowski, G., Sok, D., Crispin, M., Ward, A.B., Nemazee, D., Burton, D.R., Wilson, I.A., Zhu, J.(2018) Sci Adv 4: eaau6769-eaau6769
- PubMed: 30474059
- DOI: https://doi.org/10.1126/sciadv.aau6769
- Primary Citation of Related Structures:
6CE0 - PubMed Abstract:
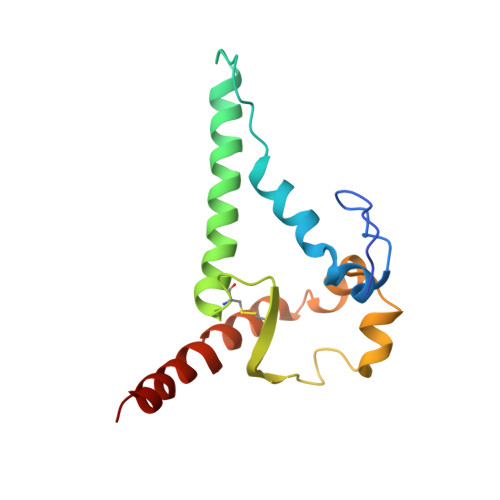
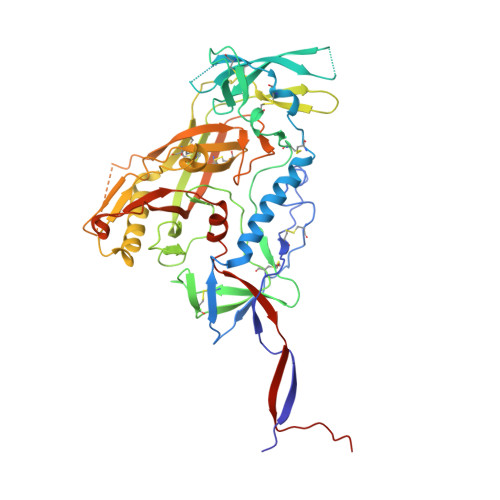
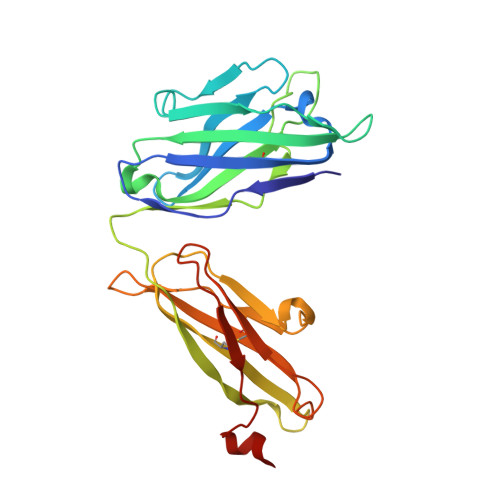
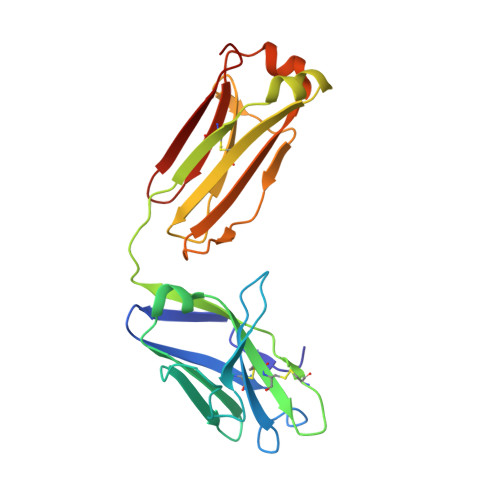
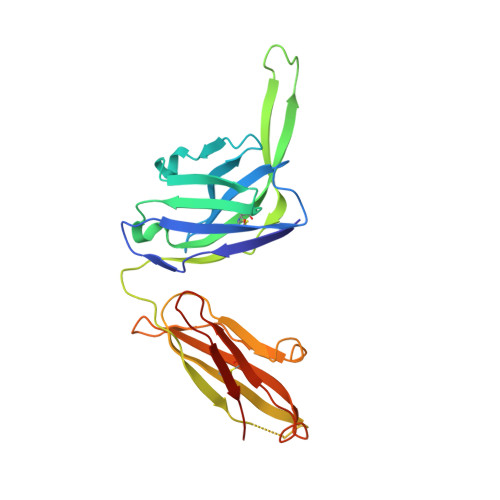
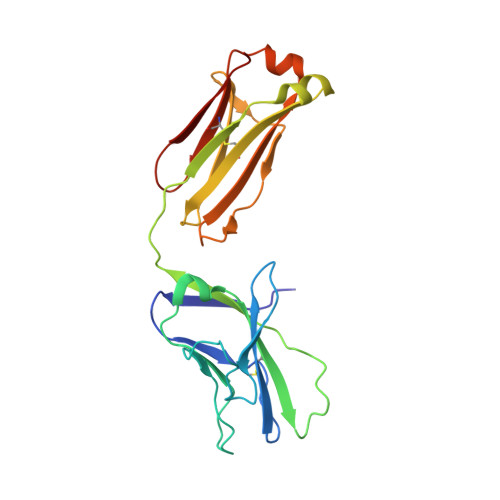
Overcoming envelope metastability is crucial to trimer-based HIV-1 vaccine design. Here, we present a coherent vaccine strategy by minimizing metastability. For 10 strains across five clades, we demonstrate that the gp41 ectodomain (gp41 ECTO ) is the main source of envelope metastability by replacing wild-type gp41 ECTO with BG505 gp41 ECTO of the uncleaved prefusion-optimized (UFO) design. These gp41 ECTO -swapped trimers can be produced in CHO cells with high yield and high purity. The crystal structure of a gp41 ECTO -swapped trimer elucidates how a neutralization-resistant tier 3 virus evades antibody recognition of the V2 apex. UFO trimers of transmitted/founder viruses and UFO trimers containing a consensus-based ancestral gp41 ECTO suggest an evolutionary root of metastability. The gp41 ECTO -stabilized trimers can be readily displayed on 24- and 60-meric nanoparticles, with incorporation of additional T cell help illustrated for a hyperstable 60-mer, I3-01. In mice and rabbits, these gp140 nanoparticles induced tier 2 neutralizing antibody responses more effectively than soluble trimers.
- Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: