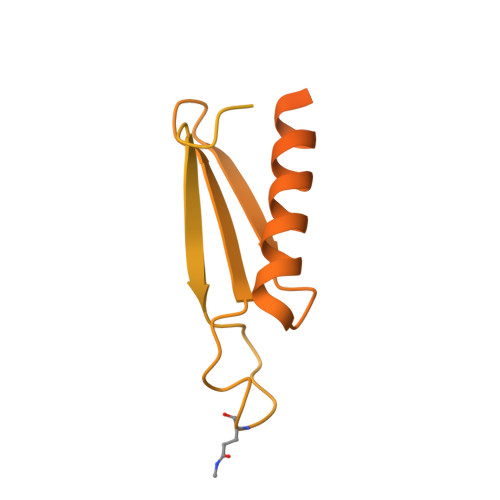
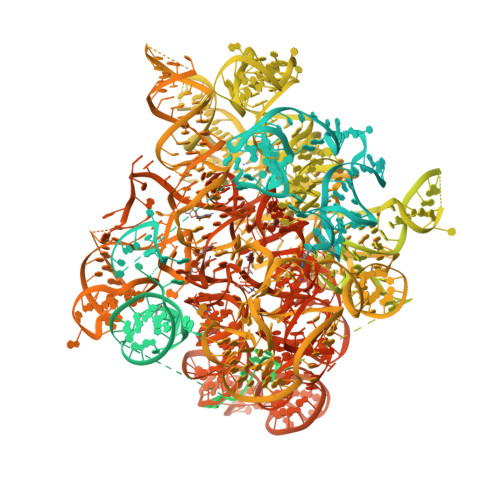
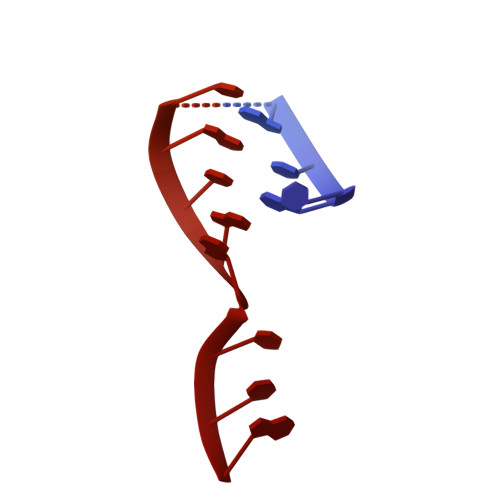
Conformation of methylated GGQ in the Peptidyl Transferase Center during Translation Termination.
Zeng, F., Jin, H.(2018) Sci Rep 8: 2349-2349
- PubMed: 29403017
- DOI: https://doi.org/10.1038/s41598-018-20107-8
- Primary Citation of Related Structures:
6C4H, 6C4I, 6C5L - PubMed Abstract:
The universally conserved Gly-Gly-Gln (GGQ) tripeptide in release factors or release factor-like surveillance proteins is required to catalyze the release of nascent peptide in the ribosome. The glutamine of the GGQ is methylated post-translationally at the N 5 position in vivo; this covalent modification is essential for optimal cell growth and efficient translation termination. However, the precise conformation of the methylated-GGQ tripeptide in the ribosome remains unknown. Using cryoEM and X-ray crystallography, we report the conformation of methylated-GGQ in the peptidyl transferase center of the ribosome during canonical translational termination and co-translation quality control. It has been suggested that the GGQ motif arose independently through convergent evolution among otherwise unrelated proteins that catalyze peptide release. The requirement for this tripeptide in the highly conserved peptidyl transferase center suggests that the conformation reported here is likely shared during termination of protein synthesis in all domains of life.
- Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, USA.
Organizational Affiliation: