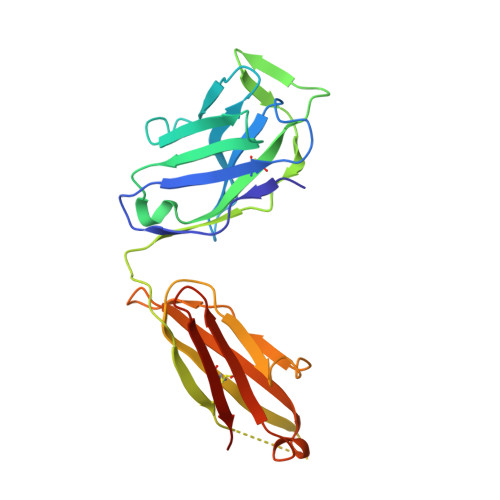
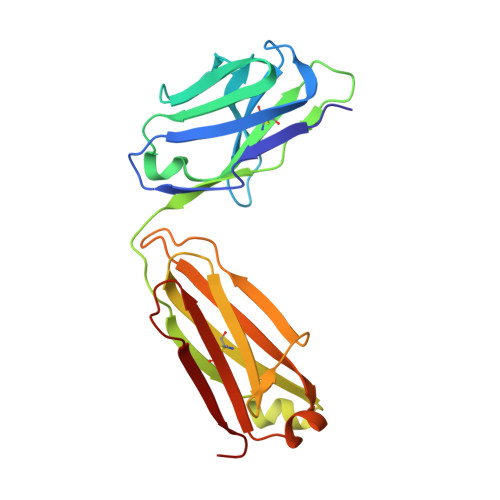
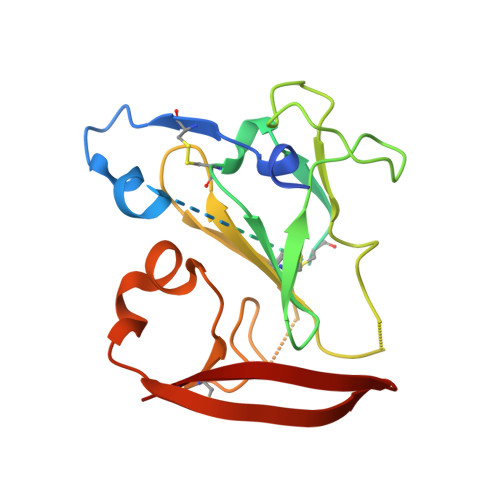
Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Tzarum, N., Giang, E., Kong, L., He, L., Prentoe, J., Augestad, E., Hua, Y., Castillo, S., Lauer, G.M., Bukh, J., Zhu, J., Wilson, I.A., Law, M.(2019) Sci Adv 5: eaav1882-eaav1882
- PubMed: 30613781
- DOI: https://doi.org/10.1126/sciadv.aav1882
- Primary Citation of Related Structures:
6BKB, 6BKC, 6BKD - PubMed Abstract:
An effective vaccine to the antigenically diverse hepatitis C virus (HCV) must target conserved immune epitopes. Here, we investigate cross-neutralization of HCV genotypes by broadly neutralizing antibodies (bNAbs) encoded by the relatively abundant human gene family V H 1-69 . We have deciphered the molecular requirements for cross-neutralization by this unique class of human antibodies from crystal structures of HCV E2 in complex with bNAbs. An unusually high binding affinity is found for germ line-reverted versions of V H 1-69 precursor antibodies, and neutralization breadth is acquired during affinity maturation. Deep sequencing analysis of an HCV-immune B cell repertoire further demonstrates the importance of the V H 1-69 gene family in the generation of HCV bNAbs. This study therefore provides critical insights into immune recognition of HCV with important implications for rational vaccine design.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: