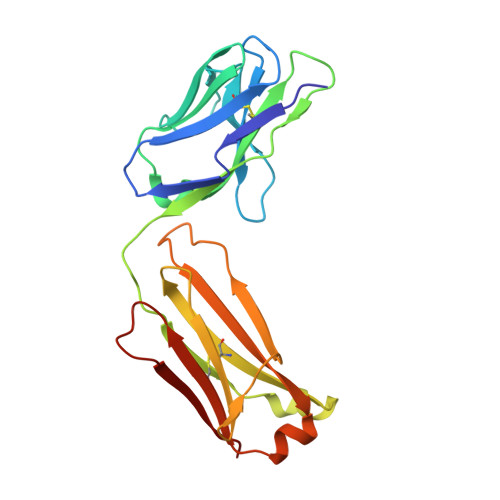
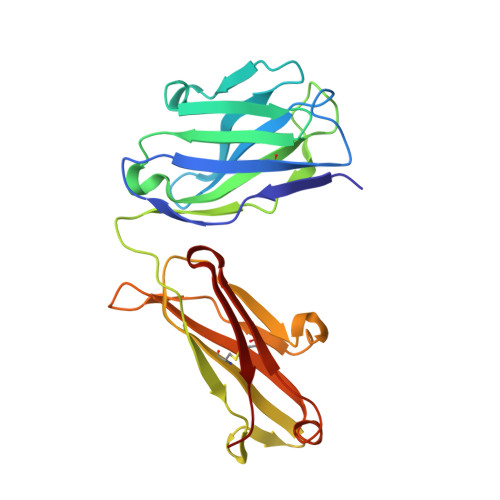
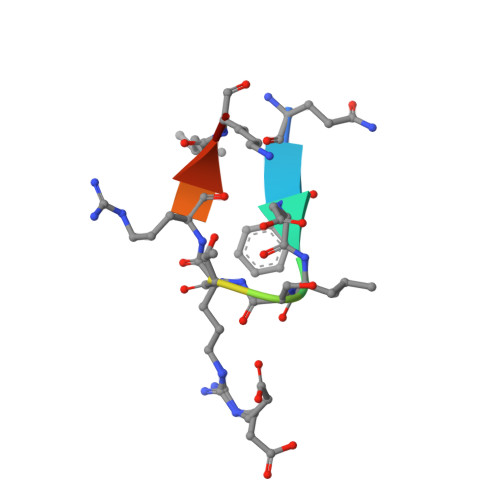
Meditope-Fab interaction: threading the hole.
Bzymek, K.P., Ma, Y., Avery, K.N., Horne, D.A., Williams, J.C.(2017) Acta Crystallogr F Struct Biol Commun 73: 688-694
- PubMed: 29199990
- DOI: https://doi.org/10.1107/S2053230X17016272
- Primary Citation of Related Structures:
6AU5, 6AXP, 6AYN, 6AZK, 6AZL - PubMed Abstract:
Meditope, a cyclic 12-residue peptide, binds to a unique binding side between the light and heavy chains of the cetuximab Fab. In an effort to improve the affinity of the interaction, it was sought to extend the side chain of Arg8 in the meditope, a residue that is accessible from the other side of the meditope binding site, in order to increase the number of interactions. These modifications included an n-butyl and n-octyl extension as well as hydroxyl, amine and carboxyl substitutions. The atomic structures of the complexes and the binding kinetics for each modified meditope indicated that each extension threaded through the Fab `hole' and that the carboxyethylarginine substitution makes a favorable interaction with the Fab, increasing the half-life of the complex by threefold compared with the unmodified meditope. Taken together, these studies provide a basis for the design of additional modifications to enhance the overall affinity of this unique interaction.
- Department of Molecular Medicine, Beckman Research Institute of City of Hope, 1710 Flower Street, Duarte, CA 91101, USA.
Organizational Affiliation: