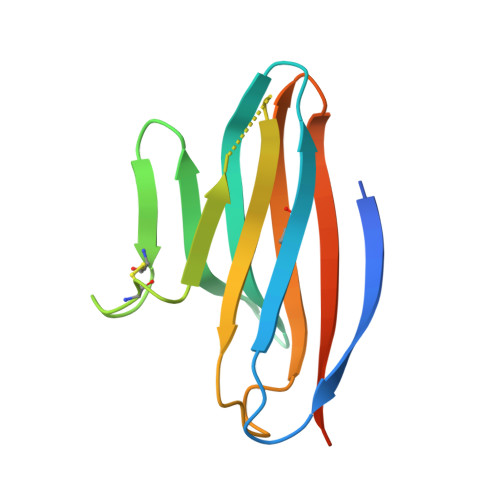
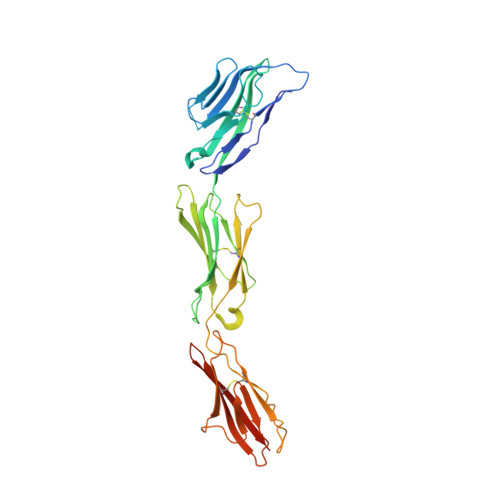
Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155.
Deuss, F.A., Watson, G.M., Fu, Z., Rossjohn, J., Berry, R.(2019) Structure 27: 219-228.e3
- PubMed: 30528596
- DOI: https://doi.org/10.1016/j.str.2018.10.023
- Primary Citation of Related Structures:
6ARQ - PubMed Abstract:
CD96, DNAM-1, and TIGIT constitute a group of immunoglobulin superfamily receptors that are key regulators of tumor immune surveillance. Within this axis, CD96 recognizes the adhesion molecule nectin-like protein-5 (necl-5), although the molecular basis underpinning this interaction remains unclear. We show that the first immunoglobulin domain (D1) of CD96 is sufficient to mediate a robust interaction with necl-5, but not the DNAM-1 and TIGIT ligand, nectin-2. The crystal structure of CD96-D1 bound to the necl-5 ectodomain revealed that CD96 recognized necl-5 D1 via a conserved "lock-and-key" interaction observed across TIGIT:necl complexes. Specific necl-5 recognition was underpinned by a novel structural motif within CD96, namely an "ancillary key". Mutational analysis showed that this specific residue was critical for necl-5 binding, while simultaneously providing insights into the unique ligand specificity of CD96.
- Infection and Immunity Program and The Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia; Australian Research Council Centre of Excellence in Advanced Molecular Imaging, Monash University, Clayton, VIC 3800, Australia.
Organizational Affiliation: