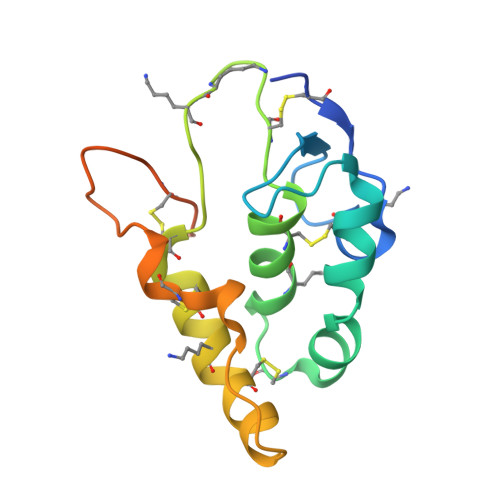
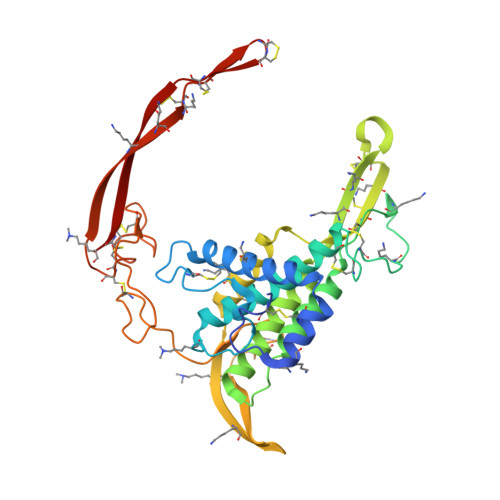
Crystal structure of a mammalian Wnt-frizzled complex.
Hirai, H., Matoba, K., Mihara, E., Arimori, T., Takagi, J.(2019) Nat Struct Mol Biol 26: 372-379
- PubMed: 31036956
- DOI: https://doi.org/10.1038/s41594-019-0216-z
- Primary Citation of Related Structures:
6AHY - PubMed Abstract:
Wnt signaling plays fundamental roles in organogenesis, tissue regeneration and cancer, but high-resolution structural information of mammalian Wnt proteins is lacking. We solved a 2.8-Å resolution crystal structure of human Wnt3 in complex with mouse Frizzled 8 Cys-rich domain (CRD). Wnt3 grabs the receptor in a manner very similar to that found in Xenopus Wnt8 complexed with the same receptor. Unlike Xenopus Wnt8-bound CRD, however, Wnt3-bound CRD formed a symmetrical dimer in the crystal by exchanging the tip of the unsaturated acyl chain attached to each Wnt3, confirming the ability of Wnt and Frizzled CRD to form a 2:2 complex. The hypervariable 'linker' region of Wnt3 formed a β-hairpin protrusion opposite from the Frizzled binding interface, consistent with its proposed role in the coreceptor recognition. Direct binding between this segment and the Wnt coreceptor LRP6 was confirmed, enabling us to build a structural model of the Wnt-Frizzled-LRP6 ternary complex.
- Laboratory of Protein Synthesis and Expression, Institute for Protein Research, Osaka University, Osaka, Japan.
Organizational Affiliation: