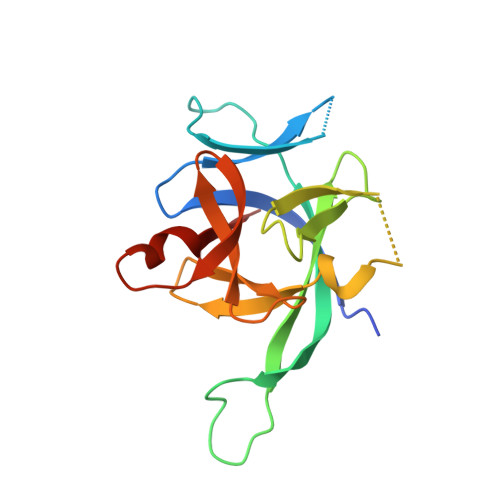
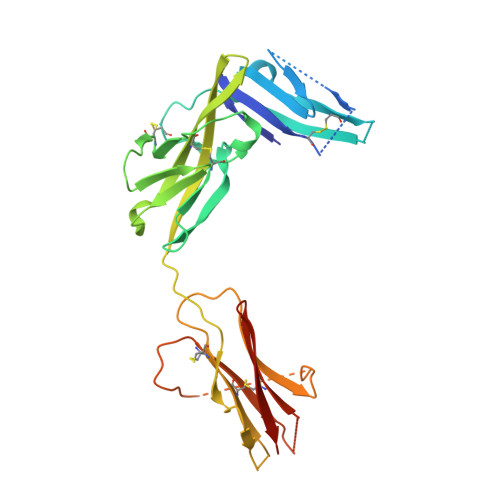
IL-1 Family Cytokines Use Distinct Molecular Mechanisms to Signal through Their Shared Co-receptor.
Gunther, S., Deredge, D., Bowers, A.L., Luchini, A., Bonsor, D.A., Beadenkopf, R., Liotta, L., Wintrode, P.L., Sundberg, E.J.(2017) Immunity 47: 510-523.e4
- PubMed: 28930661
- DOI: https://doi.org/10.1016/j.immuni.2017.08.004
- Primary Citation of Related Structures:
5VI4 - PubMed Abstract:
Within the interleukin 1 (IL-1) cytokine family, IL-1 receptor accessory protein (IL-1RAcP) is the co-receptor for eight receptor-cytokine pairs, including those involving cytokines IL-1β and IL-33. Unlike IL-1β, IL-33 does not have a signaling complex that includes both its cognate receptor, ST2, and the shared co-receptor IL-1RAcP, which we now present here. Although the IL-1β and IL-33 complexes shared structural features and engaged identical molecular surfaces of IL-1RAcP, these cytokines had starkly different strategies for co-receptor engagement and signal activation. Our data suggest that IL-1β binds to IL-1RI to properly present the cytokine to IL-1RAcP, whereas IL-33 binds to ST2 in order to conformationally constrain the cognate receptor in an IL-1RAcP-receptive state. These findings indicate that members of the IL-1 family of cytokines use distinct molecular mechanisms to signal through their shared co-receptor, and they provide the foundation from which to design new therapies to target IL-33 signaling.
- Institute of Human Virology, University of Maryland School of Medicine, Baltimore, MD 21201, USA.
Organizational Affiliation: