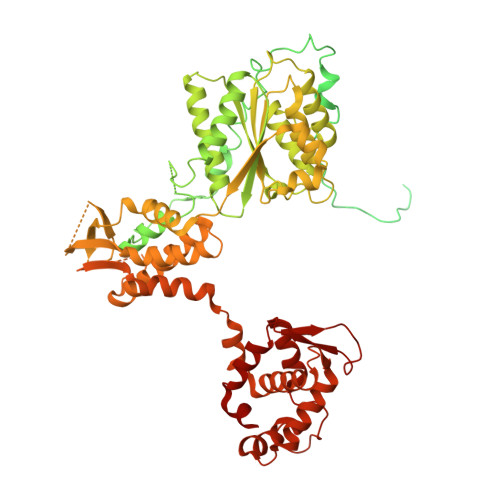
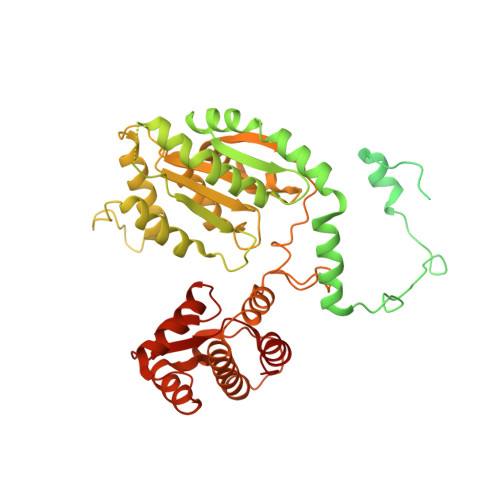
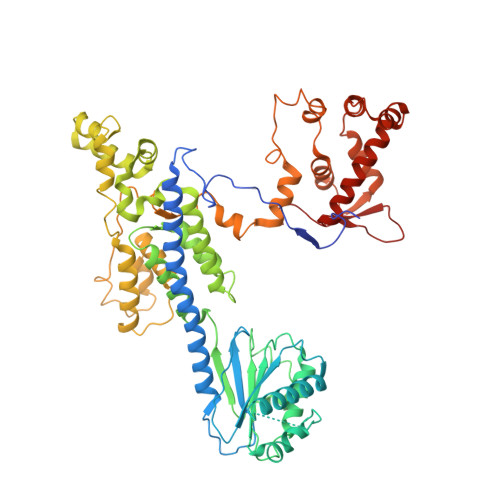
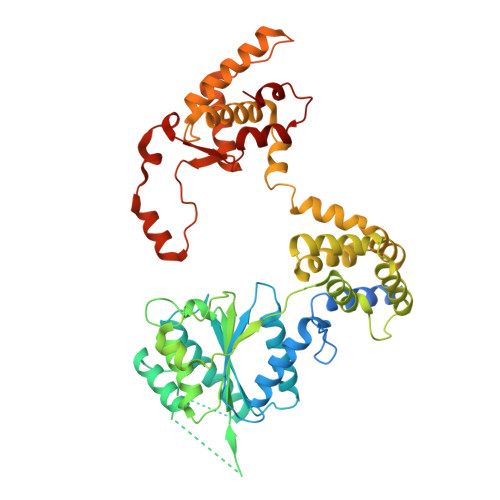
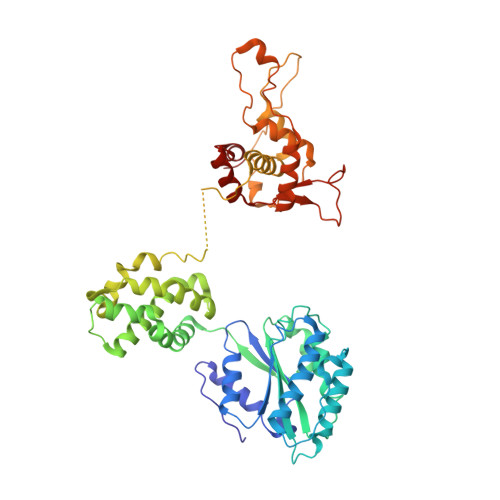
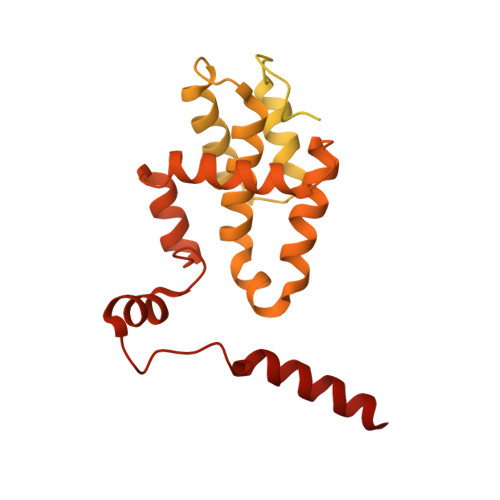
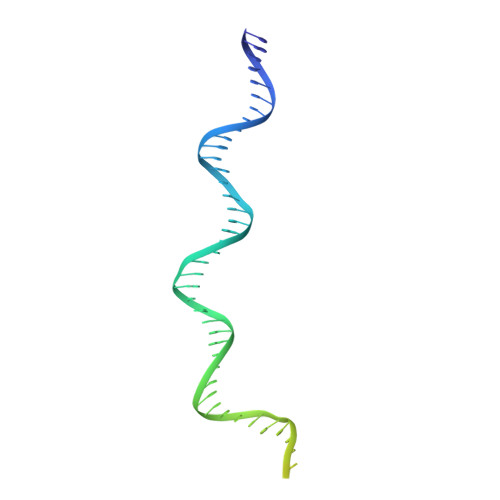
Structure of the origin recognition complex bound to DNA replication origin.
Li, N., Lam, W.H., Zhai, Y., Cheng, J., Cheng, E., Zhao, Y., Gao, N., Tye, B.K.(2018) Nature 559: 217-222
- PubMed: 29973722
- DOI: https://doi.org/10.1038/s41586-018-0293-x
- Primary Citation of Related Structures:
5ZR1 - PubMed Abstract:
The six-subunit origin recognition complex (ORC) binds to DNA to mark the site for the initiation of replication in eukaryotes. Here we report a 3 Å cryo-electron microscopy structure of the Saccharomyces cerevisiae ORC bound to a 72-base-pair origin DNA sequence that contains the ARS consensus sequence (ACS) and the B1 element. The ORC encircles DNA through extensive interactions with both phosphate backbone and bases, and bends DNA at the ACS and B1 sites. Specific recognition of thymine residues in the ACS is carried out by a conserved basic amino acid motif of Orc1 in the minor groove, and by a species-specific helical insertion motif of Orc4 in the major groove. Moreover, similar insertions into major and minor grooves are also embedded in the B1 site by basic patch motifs from Orc2 and Orc5, respectively, to contact bases and to bend DNA. This work pinpoints a conserved role of ORC in modulating DNA structure to facilitate origin selection and helicase loading in eukaryotes.
- State Key Laboratory of Membrane Biology, Peking-Tsinghua Center for Life Sciences, School of Life Sciences, Peking University, Beijing, China.
Organizational Affiliation: