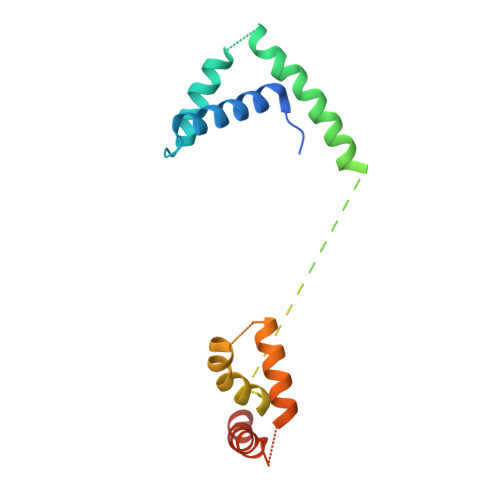Cracking the DNA Code for V(D)J Recombination
Kim, M.S., Chuenchor, W., Chen, X., Cui, Y., Zhang, X., Zhou, Z.H., Gellert, M., Yang, W.(2018) Mol Cell 70: 358-370.e4
- PubMed: 29628308
- DOI: https://doi.org/10.1016/j.molcel.2018.03.008
- Primary Citation of Related Structures:
5ZDZ, 5ZE0, 5ZE1, 5ZE2, 6CG0, 6CIJ, 6CIK, 6CIL, 6CIM - PubMed Abstract:
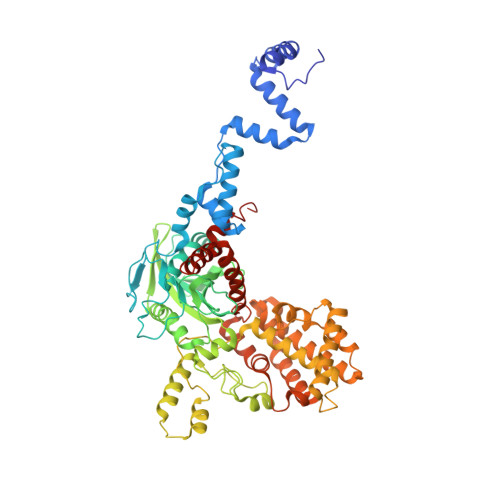
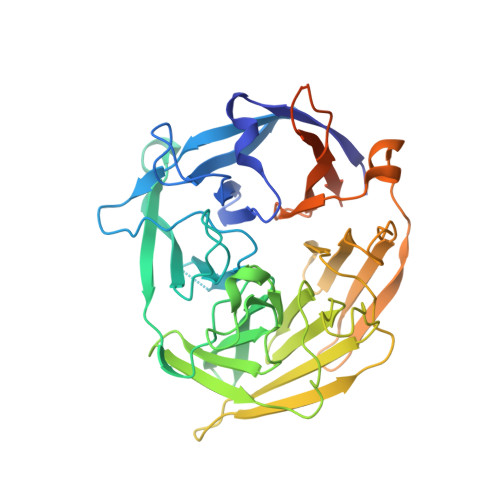
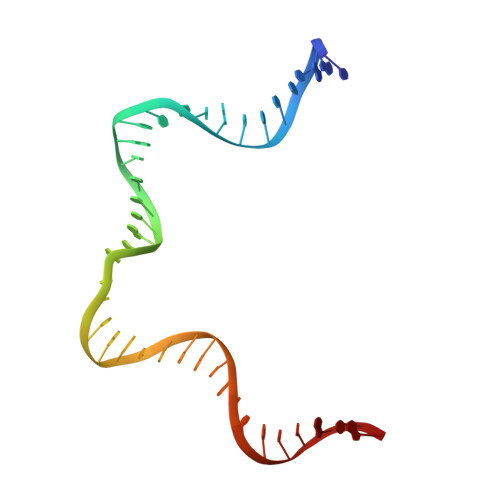
To initiate V(D)J recombination for generating the adaptive immune response of vertebrates, RAG1/2 recombinase cleaves DNA at a pair of recombination signal sequences, the 12- and 23-RSS. We have determined crystal and cryo-EM structures of RAG1/2 with DNA in the pre-reaction and hairpin-forming complexes up to 2.75 Å resolution. Both protein and DNA exhibit structural plasticity and undergo dramatic conformational changes. Coding-flank DNAs extensively rotate, shift, and deform for nicking and hairpin formation. Two intertwined RAG1 subunits crisscross four times between the asymmetric pair of severely bent 12/23-RSS DNAs. Location-sensitive bending of 60° and 150° in 12- and 23-RSS spacers, respectively, must occur for RAG1/2 to capture the nonamers and pair the heptamers for symmetric double-strand breakage. DNA pairing is thus sequence-context dependent and structure specific, which partly explains the "beyond 12/23" restriction. Finally, catalysis in crystallo reveals the process of DNA hairpin formation and its stabilization by interleaved base stacking.
- Laboratory of Molecular Biology, NIDDK, NIH, Bethesda, MD 20892, USA; Integrative Bioscience and Biotechnology, Pohang University of Science and Technology, Pohang, Gyeongbuk 37673, Republic of Korea.
Organizational Affiliation: