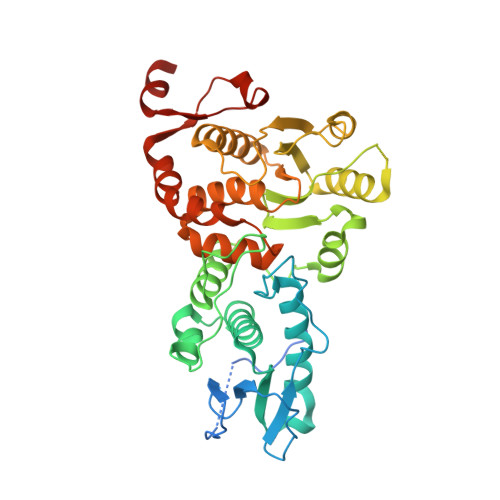
Structure and dimerization of the catalytic domain of the protein phosphatase Cdc14p, a key regulator of mitotic exit in Saccharomyces cerevisiae
Kobayashi, J., Matsuura, Y.(2017) Protein Sci 26: 2105-2112
- PubMed: 28758351
- DOI: https://doi.org/10.1002/pro.3244
- Primary Citation of Related Structures:
5XW4, 5XW5 - PubMed Abstract:
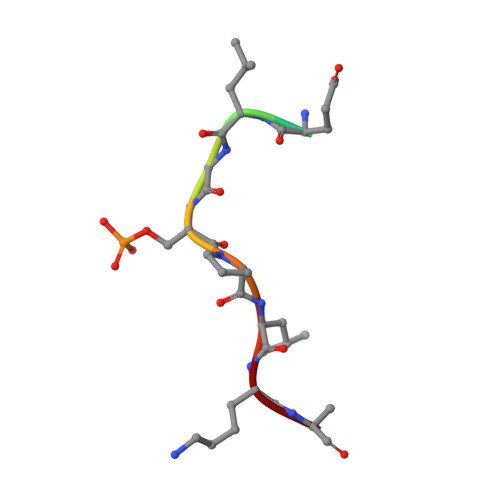
In the budding yeast Saccharomyces cerevisiae, the protein phosphatase Cdc14p orchestrates various events essential for mitotic exit. We have determined the X-ray crystal structures at 1.85 Å resolution of the catalytic domain of Cdc14p in both the apo state, and as a complex with S160-phosphorylated Swi6p peptide. Each asymmetric unit contains two Cdc14p chains arranged in an intimately associated homodimer, consistent with its oligomeric state in solution. The dimerization interface is located on the backside of the substrate-binding cleft. Structure-based mutational analyses indicate that the dimerization of Cdc14p is required for normal growth of yeast cells.
- Division of Biological Science, Nagoya University, Japan.
Organizational Affiliation: