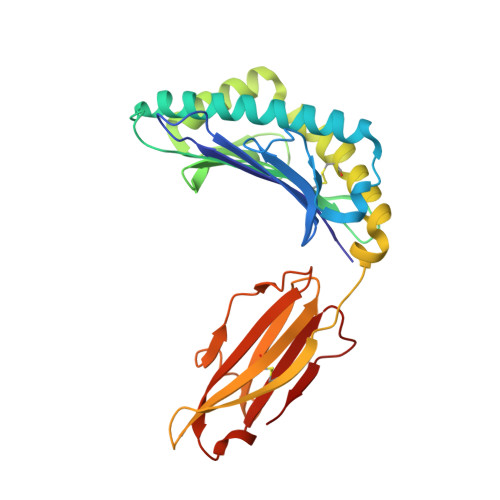
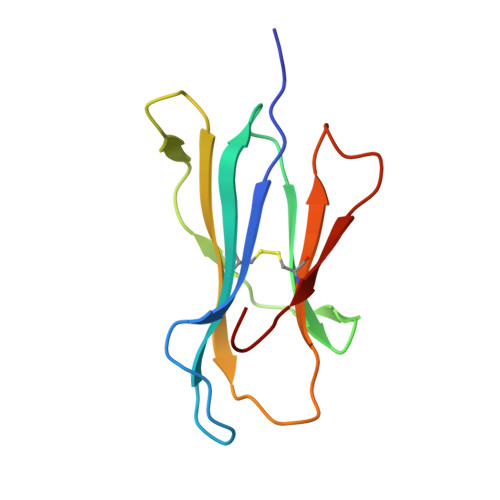
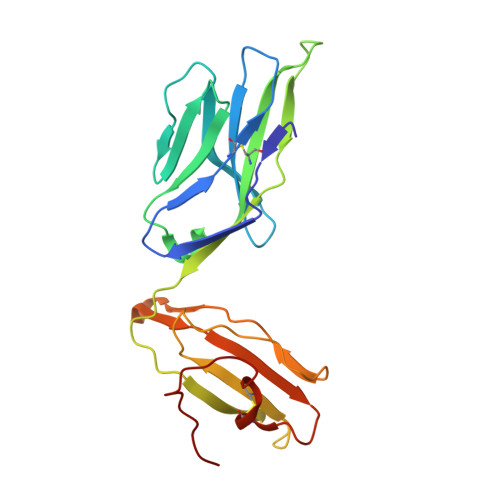
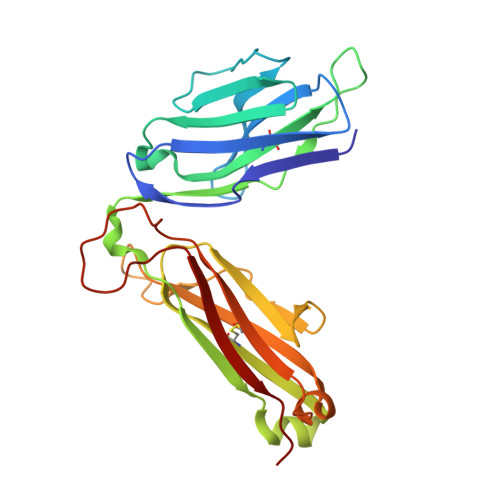
Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
Shi, Y., Kawana-Tachikawa, A., Gao, F., Qi, J., Liu, C., Gao, J., Cheng, H., Ueno, T., Iwamoto, A., Gao, G.F.(2017) J Virol 91
- PubMed: 28615212
- DOI: https://doi.org/10.1128/JVI.00725-17
- Primary Citation of Related Structures:
5XOS, 5XOT, 5XOV - PubMed Abstract:
Given the limited set of T cell receptor (TCR) V genes that are used to create TCRs that are reactive to different ligands, such as major histocompatibility complex (MHC) class I, MHC class II, and MHC-like proteins (for example, MIC molecules and CD1 molecules), the Vδ1 segment can be rearranged with Dδ-Jδ-Cδ or Jα-Cα segments to form classical γδTCRs or uncommon αβTCRs using a Vδ1 segment (δ/αβTCR). Here we have determined two complex structures of the δ/αβTCRs (S19-2 and TU55) bound to different locus-disparate MHC class I molecules with HIV peptides (HLA-A*2402-Nef138-10 and HLA-B*3501-Pol448-9). The overall binding modes resemble those of classical αβTCRs but display a strong tilt binding geometry of the Vδ1 domain toward the HLA α1 helix, due to a conserved extensive interaction between the CDR1δ loop and the N-terminal region of the α1 helix (mainly in position 62). The aromatic amino acids of the CDR1δ loop exploit different conformations ("aromatic ladder" or "aromatic hairpin") to accommodate distinct MHC helical scaffolds. This tolerance helps to explain how a particular TCR V region can similarly dock onto multiple MHC molecules and thus may potentially explain the nature of TCR cross-reactivity. In addition, the length of the CDR3δ loop could affect the extent of tilt binding of the Vδ1 domain, and adaptively, the pairing Vβ domains adjust their mass centers to generate differential MHC contacts, hence probably ensuring TCR specificity for a certain peptide-MHC class I (pMHC-I). Our data have provided further structural insights into the TCR recognition of classical pMHC-I molecules, unifying cross-reactivity and specificity. IMPORTANCE The specificity of αβ T cell recognition is determined by the CDR loops of the αβTCR, and the general mode of binding of αβTCRs to pMHC has been established over the last decade. Due to the intrinsic genomic structure of the TCR α/δ chain locus, some Vδ segments can rearrange with the Cα segment, forming a hybrid VδCαVβCβ TCR, the δ/αβTCR. However, the basis for the molecular recognition of such TCRs of their ligands is elusive. Here an αβTCR using the Vδ1 segment, S19-2, was isolated from an HIV-infected patient in an HLA-A*24:02-restricted manner. We then solved the crystal structures of the S19-2 TCR and another δ/αβTCR, TU55, bound to their respective ligands, revealing a conserved Vδ1 binding feature. Further binding kinetics analysis revealed that the S19-2 and TU55 TCRs bind pHLA very tightly and in a long-lasting manner. Our results illustrate the mode of binding of a TCR using the Vδ1 segment to its ligand, virus-derived pHLA.
- CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: