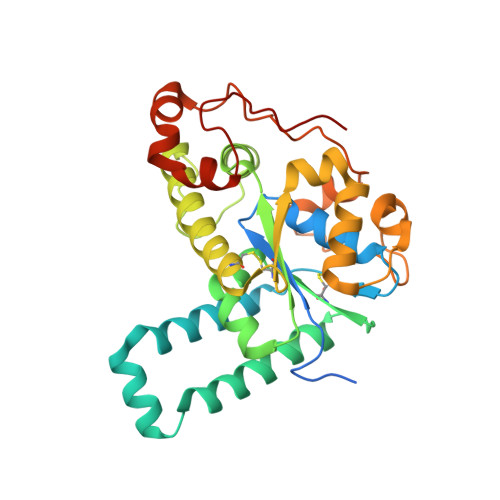
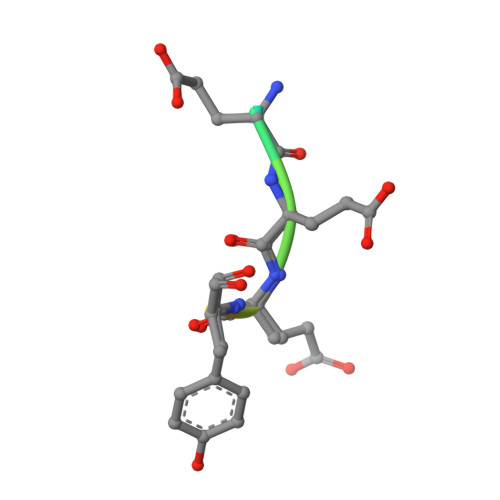
Structural basis for the broad substrate specificity of the human tyrosylprotein sulfotransferase-1.
Tanaka, S., Nishiyori, T., Kojo, H., Otsubo, R., Tsuruta, M., Kurogi, K., Liu, M.C., Suiko, M., Sakakibara, Y., Kakuta, Y.(2017) Sci Rep 7: 8776-8776
- PubMed: 28821720
- DOI: https://doi.org/10.1038/s41598-017-07141-8
- Primary Citation of Related Structures:
5WRI, 5WRJ - PubMed Abstract:
Tyrosylprotein sulfotransferases (TPSTs) are enzymes that catalyze post-translational tyrosine sulfation of proteins. In humans, there are only two TPST isoforms, designated TPST1 and TPST2. In a previous study, we reported the crystal structure of TPST2, which revealed the catalytic mechanism of the tyrosine sulfation reaction. However, detailed molecular mechanisms underlying how TPSTs catalyse a variety of substrate proteins with different efficiencies and how TPSTs catalyze the sulfation of multiple tyrosine residues in a substrate protein remain unresolved. Here, we report two crystal structures of the human TPST1 complexed with two substrate peptides that are catalysed by human TPST1 with significantly different efficiencies. The distinct binding modes found in the two complexes provide insight into the sulfation mechanism for these substrates. The present study provides valuable information describing the molecular mechanism of post-translational protein modifications catalysed by TPSTs.
- Department of Bioscience and Biotechnology, Faculty of Agriculture, Kyushu University, Hakozaki 6-10-1, Fukuoka, 812-8581, Japan.
Organizational Affiliation: