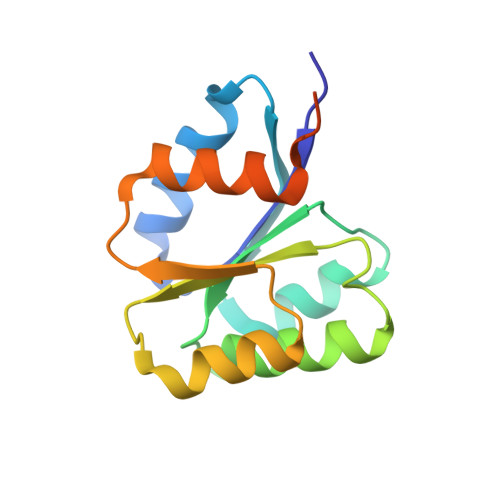
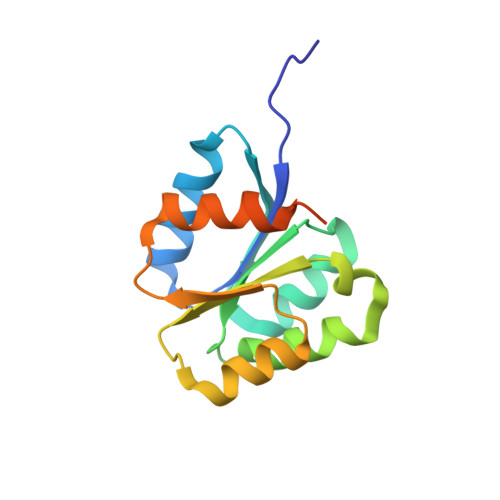
Crystal structure of the inactive state of the receiver domain of Spo0A from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
Lee, C.W., Park, S.H., Lee, S.G., Shin, S.C., Han, S.J., Kim, H.W., Park, H.H., Kim, S., Kim, H.J., Park, H., Park, H., Lee, J.H.(2017) J Microbiol 55: 464-474
- PubMed: 28281198
- DOI: https://doi.org/10.1007/s12275-017-6599-9
- Primary Citation of Related Structures:
5WQ0 - PubMed Abstract:
The two-component phosphorelay system is the most prevalent mechanism for sensing and transducing environmental signals in bacteria. Spore formation, which relies on the two-component phosphorelay system, enables the long-term survival of the glacial bacterium Paenisporosarcina sp. TG-14 in the extreme cold environment. Spo0A is a key response regulator of the phosphorelay system in the early stage of spore formation. The protein is composed of a regulatory N-terminal phospho-receiver domain and a DNA-binding C-terminal activator domain. We solved the three-dimensional structure of the unphosphorylated (inactive) form of the receiver domain of Spo0A (PaSpo0A-R) from Paenisporosarcina sp. TG-14. A structural comparison with phosphorylated (active form) Spo0A from Bacillus stearothermophilus (BsSpo0A) showed minor notable differences. A molecular dynamics study of a model of the active form and the crystal structures revealed significant differences in the α4 helix and the preceding loop region where phosphorylation occurs. Although an oligomerization study of PaSpo0A-R by analytical ultracentrifugation (AUC) has shown that the protein is in a monomeric state in solution, both crosslinking and crystal-packing analyses indicate the possibility of weak dimer formation by a previously undocumented mechanism. Collectively, these observations provide insight into the mechanism of phosphorylation-dependent activation unique to Spo0A.
- Unit of Polar Genomics, Korea Polar Research Institute, Incheon, 21990, Republic of Korea.
Organizational Affiliation: