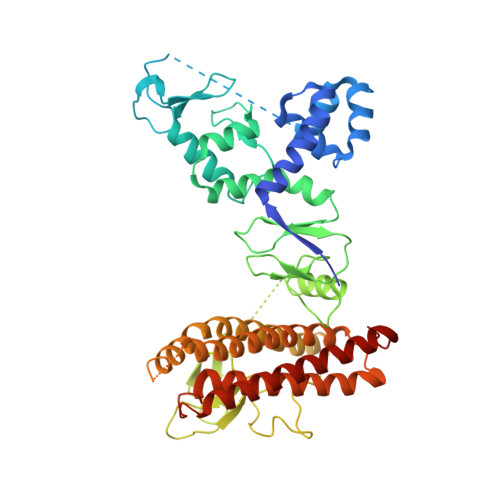
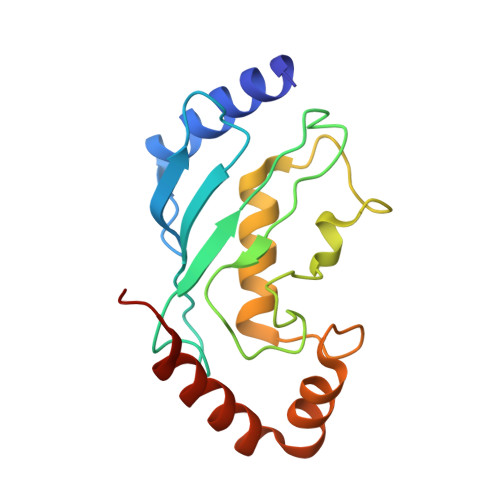
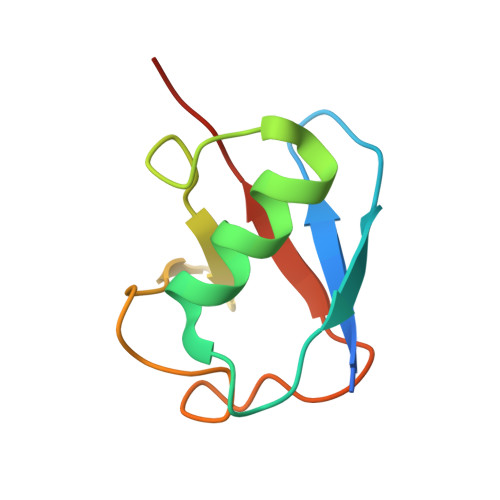
Structural Studies of HHARI/UbcH7Ub Reveal Unique E2Ub Conformational Restriction by RBR RING1.
Dove, K.K., Olszewski, J.L., Martino, L., Duda, D.M., Wu, X.S., Miller, D.J., Reiter, K.H., Rittinger, K., Schulman, B.A., Klevit, R.E.(2017) Structure 25: 890-900.e5
- PubMed: 28552575
- DOI: https://doi.org/10.1016/j.str.2017.04.013
- Primary Citation of Related Structures:
5UDH - PubMed Abstract:
RING-between-RING (RBR) E3s contain RING1 domains that are structurally similar yet mechanistically distinct from canonical RING domains. Both types of E3 bind E2∼ubiquitin (E2∼Ub) via their RINGs but canonical RING E3s promote closed E2∼Ub conformations required for direct Ub transfer from the E2 to substrate, while RBR RING1s promote open E2∼Ub to favor Ub transfer to the E3 active site. This different RING/E2∼Ub conformation determines its direct target, which for canonical RING E3s is typically a substrate or substrate-linked Ub, but is the E3 active-site cysteine in the case of RBR-type E3s. Here we show that a short extension of HHARI RING1, namely Zn 2+ -loop II, not present in any RING E3s, acts as a steric wedge to disrupt closed E2∼Ub, providing a structural explanation for the distinctive RING1-dependent conformational restriction mechanism utilized by RBR E3s.
- Department of Biochemistry, University of Washington, 1705 Northeast Pacific Street, Seattle, WA 98195, USA.
Organizational Affiliation: