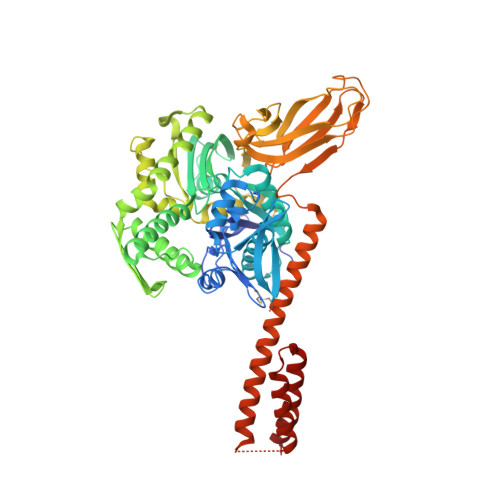
Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain.
Gumiero, A., Conz, C., Gese, G.V., Zhang, Y., Weyer, F.A., Lapouge, K., Kappes, J., von Plehwe, U., Schermann, G., Fitzke, E., Wolfle, T., Fischer, T., Rospert, S., Sinning, I.(2016) Nat Commun 7: 13563-13563
- PubMed: 27882919
- DOI: https://doi.org/10.1038/ncomms13563
- Primary Citation of Related Structures:
5TKY - PubMed Abstract:
Cotranslational chaperones assist in de novo folding of nascent polypeptides in all organisms. In yeast, the heterodimeric ribosome-associated complex (RAC) forms a unique chaperone triad with the Hsp70 homologue Ssb. We report the X-ray structure of full length Ssb in the ATP-bound open conformation at 2.6 Å resolution and identify a positively charged region in the α-helical lid domain (SBDα), which is present in all members of the Ssb-subfamily of Hsp70s. Mutational analysis demonstrates that this region is strictly required for ribosome binding. Crosslinking shows that Ssb binds close to the tunnel exit via contacts with both, ribosomal proteins and rRNA, and that specific contacts can be correlated with switching between the open (ATP-bound) and closed (ADP-bound) conformation. Taken together, our data reveal how Ssb dynamics on the ribosome allows for the efficient interaction with nascent chains upon RAC-mediated activation of ATP hydrolysis.
- Heidelberg University Biochemistry Center (BZH), INF 328, D-69120 Heidelberg, Germany.
Organizational Affiliation: