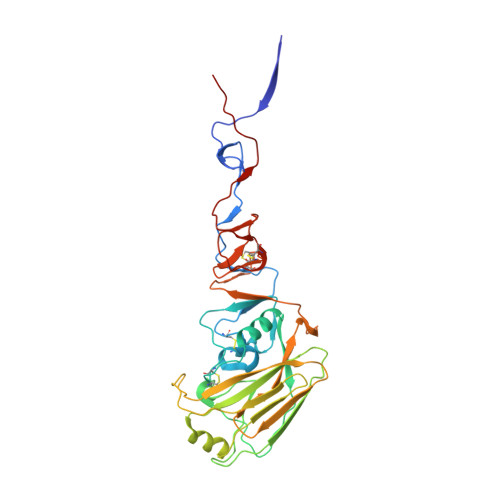
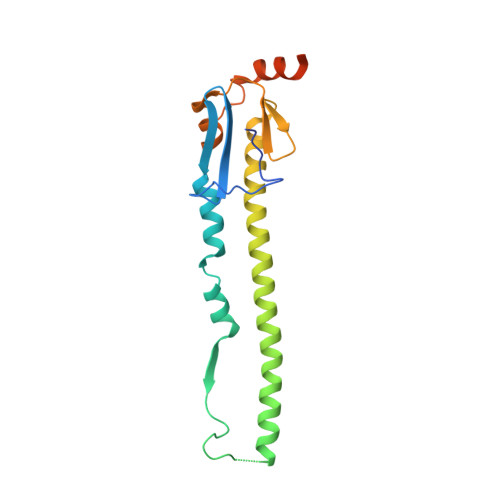
Unique Structural Features of Influenza Virus H15 Hemagglutinin.
Tzarum, N., McBride, R., Nycholat, C.M., Peng, W., Paulson, J.C., Wilson, I.A.(2017) J Virol 91
- PubMed: 28404848
- DOI: https://doi.org/10.1128/JVI.00046-17
- Primary Citation of Related Structures:
5TG8, 5TG9 - PubMed Abstract:
Influenza A H15 viruses are members of a subgroup (H7-H10-H15) of group 2 hemagglutinin (HA) subtypes that include H7N9 and H10N8 viruses that were isolated from humans during 2013. The isolation of avian H15 viruses is, however, quite rare and, until recently, geographically restricted to wild shorebirds and waterfowl in Australia. The HAs of H15 viruses contain an insertion in the 150-loop (loop beginning at position 150) of the receptor-binding site common to this subgroup and a unique insertion in the 260-loop compared to any other subtype. Here, we show that the H15 HA has a high preference for avian receptor analogs by glycan array analyses. The H15 HA crystal structure reveals that it is structurally closest to H7N9 HA, but the head domain of the H15 trimer is wider than all other HAs due to a tilt and opening of the HA1 subunits of the head domain. The extended 150-loop of the H15 HA retains the conserved conformation as in H7 and H10 HAs. Furthermore, the elongated 260-loop increases the exposed HA surface and can contribute to antigenic variation in H15 HAs. Since avian-origin H15 HA viruses have been shown to cause enhanced disease in mammalian models, further characterization and immune surveillance of H15 viruses are warranted. IMPORTANCE In the last 2 decades, an apparent increase has been reported for cases of human infection by emerging avian influenza A virus subtypes, including H7N9 and H10N8 viruses isolated during 2013. H15 is the other member of the subgroup of influenza A virus group 2 hemagglutinins (HAs) that also include H7 and H10. H15 viruses have been restricted to Australia, but recent isolation of H15 viruses in western Siberia suggests that they could be spread more globally via the avian flyways that converge and emanate from this region. Here we report on characterization of the three-dimensional structure and receptor specificity of the H15 hemagglutinin, revealing distinct features and specificities that can aid in global surveillance of such viruses for potential spread and emerging threat to the human population.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, California, USA.
Organizational Affiliation: