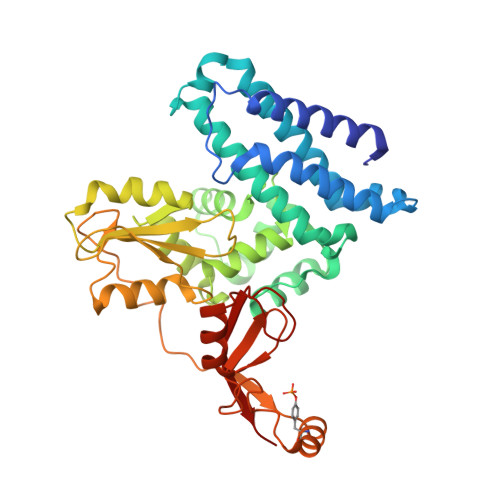
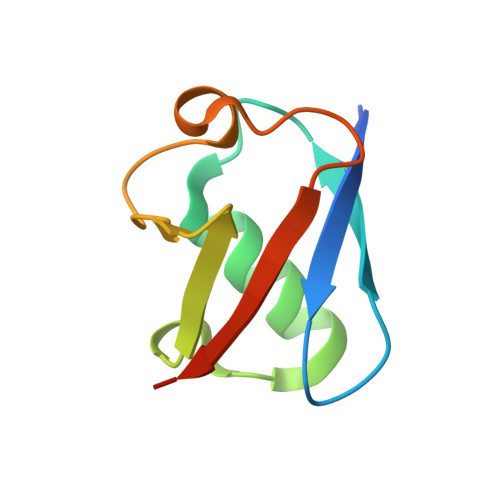
A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Gabrielsen, M., Buetow, L., Nakasone, M.A., Ahmed, S.F., Sibbet, G.J., Smith, B.O., Zhang, W., Sidhu, S.S., Huang, D.T.(2017) Mol Cell 68: 456-470.e10
- PubMed: 29053960
- DOI: https://doi.org/10.1016/j.molcel.2017.09.027
- Primary Citation of Related Structures:
5O6S, 5O6T, 5O75, 5O76 - PubMed Abstract:
RING and U-box E3 ubiquitin ligases regulate diverse eukaryotic processes and have been implicated in numerous diseases, but targeting these enzymes remains a major challenge. We report the development of three ubiquitin variants (UbVs), each binding selectively to the RING or U-box domain of a distinct E3 ligase: monomeric UBE4B, phosphorylated active CBL, or dimeric XIAP. Structural and biochemical analyses revealed that UbVs specifically inhibited the activity of UBE4B or phosphorylated CBL by blocking the E2∼Ub binding site. Surprisingly, the UbV selective for dimeric XIAP formed a dimer to stimulate E3 activity by stabilizing the closed E2∼Ub conformation. We further verified the inhibitory and stimulatory functions of UbVs in cells. Our work provides a general strategy to inhibit or activate RING/U-box E3 ligases and provides a resource for the research community to modulate these enzymes.
- Cancer Research UK Beatson Institute, Garscube Estate, Switchback Road, Glasgow G61 1BD, UK.
Organizational Affiliation: