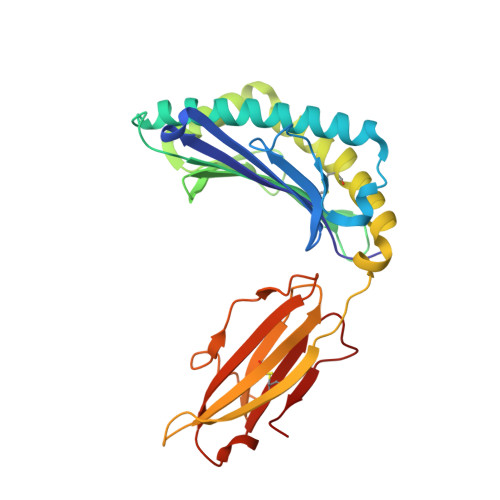
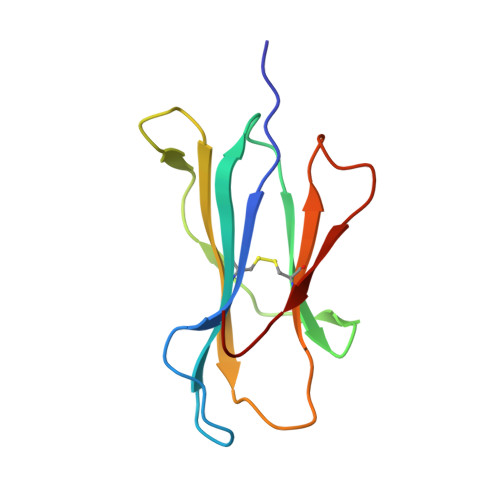
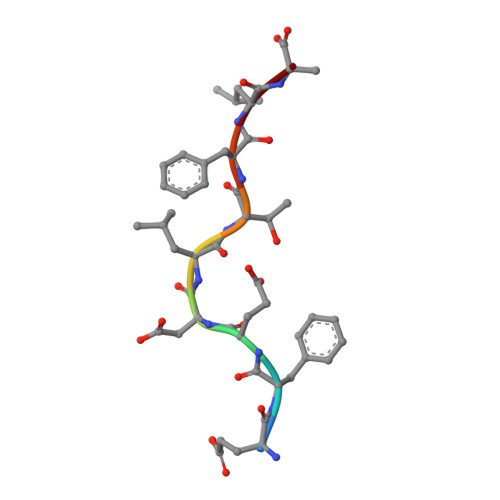
Induction of influenza-specific local CD8 T-cells in the respiratory tract after aerosol delivery of vaccine antigen or virus in the Babraham inbred pig.
Tungatt, K., Dolton, G., Morgan, S.B., Attaf, M., Fuller, A., Whalley, T., Hemmink, J.D., Porter, E., Szomolay, B., Montoya, M., Hammond, J.A., Miles, J.J., Cole, D.K., Townsend, A., Bailey, M., Rizkallah, P.J., Charleston, B., Tchilian, E., Sewell, A.K.(2018) PLoS Pathog 14: e1007017-e1007017
- PubMed: 29772011
- DOI: https://doi.org/10.1371/journal.ppat.1007017
- Primary Citation of Related Structures:
5NPZ, 5NQ0, 5NQ1, 5NQ2, 5NQ3 - PubMed Abstract:
There is increasing evidence that induction of local immune responses is a key component of effective vaccines. For respiratory pathogens, for example tuberculosis and influenza, aerosol delivery is being actively explored as a method to administer vaccine antigens. Current animal models used to study respiratory pathogens suffer from anatomical disparity with humans. The pig is a natural and important host of influenza viruses and is physiologically more comparable to humans than other animal models in terms of size, respiratory tract biology and volume. It may also be an important vector in the birds to human infection cycle. A major drawback of the current pig model is the inability to analyze antigen-specific CD8+ T-cell responses, which are critical to respiratory immunity. Here we address this knowledge gap using an established in-bred pig model with a high degree of genetic identity between individuals, including the MHC (Swine Leukocyte Antigen (SLA)) locus. We developed a toolset that included long-term in vitro pig T-cell culture and cloning and identification of novel immunodominant influenza-derived T-cell epitopes. We also generated structures of the two SLA class I molecules found in these animals presenting the immunodominant epitopes. These structures allowed definition of the primary anchor points for epitopes in the SLA binding groove and established SLA binding motifs that were used to successfully predict other influenza-derived peptide sequences capable of stimulating T-cells. Peptide-SLA tetramers were constructed and used to track influenza-specific T-cells ex vivo in blood, the lungs and draining lymph nodes. Aerosol immunization with attenuated single cycle influenza viruses (S-FLU) induced large numbers of CD8+ T-cells specific for conserved NP peptides in the respiratory tract. Collectively, these data substantially increase the utility of pigs as an effective model for studying protective local cellular immunity against respiratory pathogens.
- Division of Infection and Immunity, Cardiff University School of Medicine, Cardiff, Wales, United Kingdom.
Organizational Affiliation: