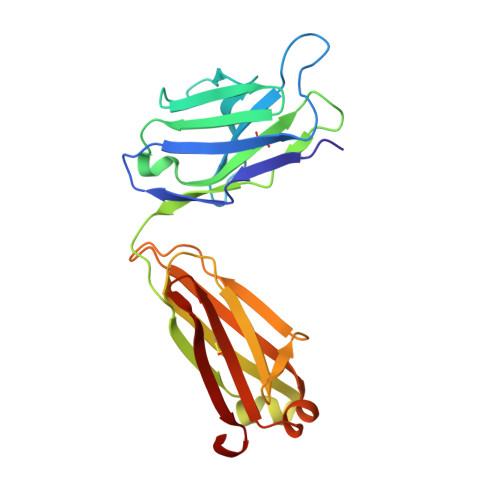
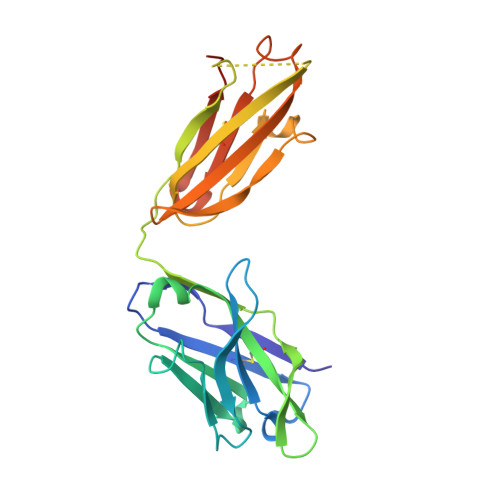
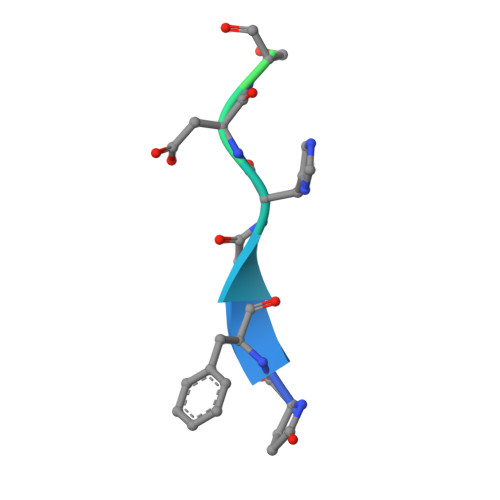
Structural and functional analyses of pyroglutamate-amyloid-beta-specific antibodies as a basis for Alzheimer immunotherapy.
Piechotta, A., Parthier, C., Kleinschmidt, M., Gnoth, K., Pillot, T., Lues, I., Demuth, H.U., Schilling, S., Rahfeld, J.U., Stubbs, M.T.(2017) J Biological Chem 292: 12713-12724
- PubMed: 28623233
- DOI: https://doi.org/10.1074/jbc.M117.777839
- Primary Citation of Related Structures:
5MY4, 5MYK, 5MYO, 5MYX - PubMed Abstract:
Alzheimer disease is associated with deposition of the amyloidogenic peptide Aβ in the brain. Passive immunization using Aβ-specific antibodies has been demonstrated to reduce amyloid deposition both in vitro and in vivo Because N-terminally truncated pyroglutamate (pE)-modified Aβ species (Aβ pE3 ) exhibit enhanced aggregation potential and propensity to form toxic oligomers, they represent particularly attractive targets for antibody therapy. Here we present three separate monoclonal antibodies that specifically recognize Aβ pE3 with affinities of 1-10 nm and inhibit Aβ pE3 fibril formation in vitro. In vivo application of one of these resulted in improved memory in Aβ pE3 oligomer-treated mice. Crystal structures of F ab -Aβ pE3 complexes revealed two distinct binding modes for the peptide. Juxtaposition of pyroglutamate pE3 and the F4 side chain (the "pEF head") confers a pronounced bulky hydrophobic nature to the Aβ pE3 N terminus that might explain the enhanced aggregation properties of the modified peptide. The deep burial of the pEF head by two of the antibodies explains their high target specificity and low cross-reactivity, making them promising candidates for the development of clinical antibodies.
- Probiodrug AG, Weinbergweg 22, 06120 Halle (Saale), Germany; Institute of Biotechnology, Martin Luther University, 06108 Halle-Wittenberg, Germany; Department of Molecular Drug Biochemistry and Therapy, Fraunhofer Institute for Cell Therapy and Immunology, Weinbergweg 22, 06120 Halle, Germany.
Organizational Affiliation: