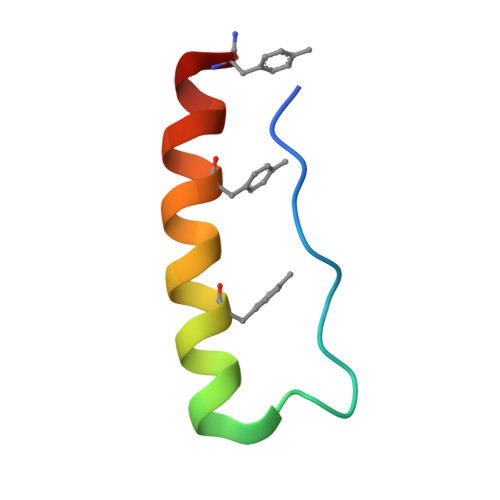
Engineering protein stability with atomic precision in a monomeric miniprotein.
Baker, E.G., Williams, C., Hudson, K.L., Bartlett, G.J., Heal, J.W., Porter Goff, K.L., Sessions, R.B., Crump, M.P., Woolfson, D.N.(2017) Nat Chem Biol 13: 764-770
- PubMed: 28530710
- DOI: https://doi.org/10.1038/nchembio.2380
- Primary Citation of Related Structures:
5LO2, 5LO3, 5LO4 - PubMed Abstract:
Miniproteins simplify the protein-folding problem, allowing the dissection of forces that stabilize protein structures. Here we describe PPα-Tyr, a designed peptide comprising an α-helix buttressed by a polyproline II helix. PPα-Tyr is water soluble and monomeric, and it unfolds cooperatively with a midpoint unfolding temperature (T M ) of 39 °C. NMR structures of PPα-Tyr reveal proline residues docked between tyrosine side chains, as designed. The stability of PPα is sensitive to modifications in the aromatic residues: replacing tyrosine with phenylalanine, i.e., changing three solvent-exposed hydroxyl groups to protons, reduces the T M to 20 °C. We attribute this result to the loss of CH-π interactions between the aromatic and proline rings, which we probe by substituting the aromatic residues with nonproteinogenic side chains. In analyses of natural protein structures, we find a preference for proline-tyrosine interactions over other proline-containing pairs, and observe abundant CH-π interactions in biologically important complexes between proline-rich ligands and SH3 and similar domains.
- School of Chemistry, University of Bristol, Bristol, UK.
Organizational Affiliation: