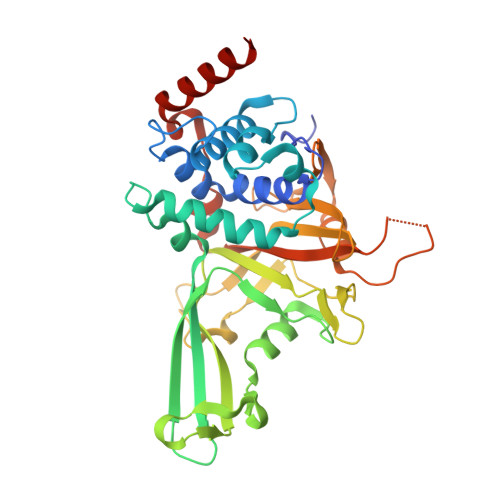
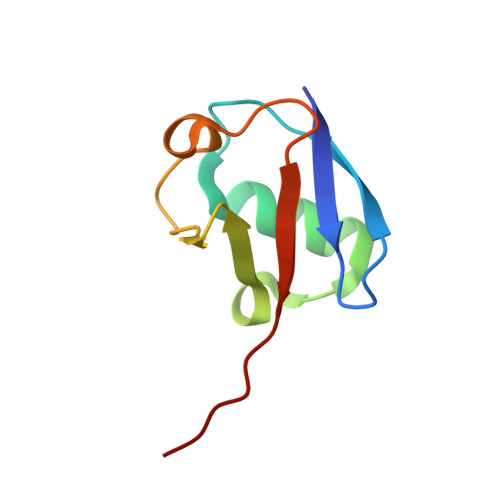
Selectively Modulating Conformational States of USP7 Catalytic Domain for Activation.
Ozen, A., Rouge, L., Bashore, C., Hearn, B.R., Skelton, N.J., Dueber, E.C.(2018) Structure 26: 72-84.e7
- PubMed: 29249604
- DOI: https://doi.org/10.1016/j.str.2017.11.010
- Primary Citation of Related Structures:
5KYB, 5KYC, 5KYD, 5KYE, 5KYF - PubMed Abstract:
Ubiquitin-specific protease 7 (USP7) deubiquitinase activity is controlled by a number of regulatory factors, including stimulation by intramolecular accessory domains. Alone, the USP7 catalytic domain (USP7cd) shows limited activity and apo USP7cd crystal structures reveal a disrupted catalytic triad. By contrast, ubiquitin-conjugated USP7cd structures demonstrate the canonical cysteine protease active-site geometry; however, the structural features of the USP7cd that stabilize the inactive conformation and the mechanism of transition between inactive and active states remain unclear. Here we use comparative structural analyses, molecular dynamics simulations, and in silico sequence re-engineering via directed sampling by RosettaDesign to identify key molecular determinants of USP7cd activation and successfully engineer USP7cd for improved activity. Full kinetic analysis and multiple X-ray crystal structures of our designs indicate that electrostatic interactions in the distal "switching loop" region and local packing in the hydrophobic core mediate subtle but significant conformational changes that modulate USP7cd activation.
- Department of Early Discovery Biochemistry, Genentech Inc., South San Francisco, CA 94080, USA; Department of Discovery Chemistry, Genentech Inc., South San Francisco, CA 94080, USA.
Organizational Affiliation: