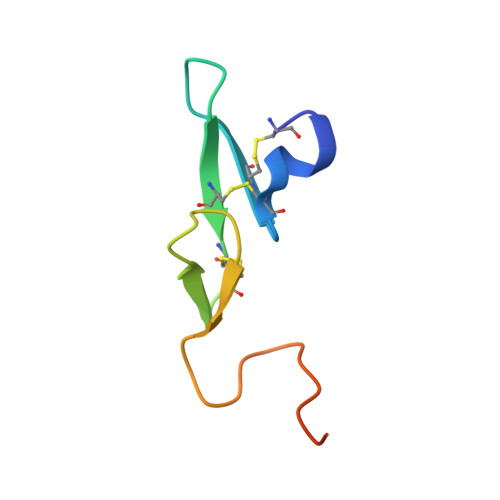
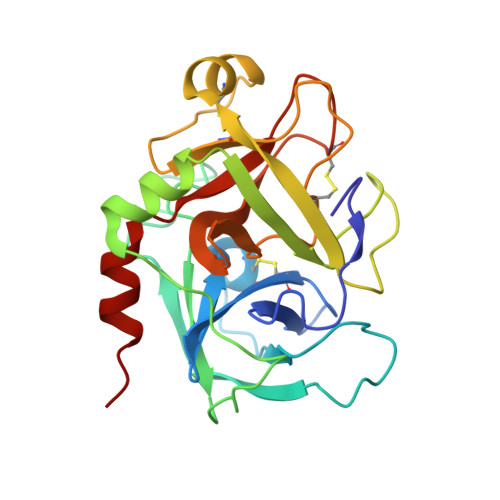
Releasing the brakes in coagulation Factor IXa by co-operative maturation of the substrate-binding site.
Kristensen, L.H., Olsen, O.H., Blouse, G.E., Brandstetter, H.(2016) Biochem J 473: 2395-2411
- PubMed: 27208168
- DOI: https://doi.org/10.1042/BCJ20160336
- Primary Citation of Related Structures:
5JB8, 5JB9, 5JBA, 5JBB, 5JBC - PubMed Abstract:
Coagulation Factor IX is positioned at the merging point of the intrinsic and extrinsic blood coagulation cascades. Factor IXa (activated Factor IX) serves as the trigger for amplification of coagulation through formation of the so-called Xase complex, which is a ternary complex of Factor IXa, its substrate Factor X and the cofactor Factor VIIIa on the surface of activated platelets. Within the Xase complex the substrate turnover by Factor IXa is enhanced 200000-fold; however, the mechanistic and structural basis for this dramatic enhancement remains only partly understood. A multifaceted approach using enzymatic, biophysical and crystallographic methods to evaluate a key set of activity-enhanced Factor IXa variants has demonstrated a delicately balanced bidirectional network. Essential molecular interactions across multiple regions of the Factor IXa molecule co-operate in the maturation of the active site. This maturation is specifically facilitated by long-range communication through the Ile(212)-Ile(213) motif unique to Factor IXa and a flexibility of the 170-loop that is further dependent on the conformation in the Cys(168)-Cys(182) disulfide bond. Ultimately, the network consists of compensatory brakes (Val(16) and Ile(213)) and accelerators (Tyr(99) and Phe(174)) that together allow for a subtle fine-tuning of enzymatic activity.
- Haemophilia Biochemistry, Novo Nordisk A/S, Novo Nordisk Park 1, 2760 Måløv, Denmark Structural Biology Group, Department of Molecular Biology, University of Salzburg, Billrothstrasse 11, 5020 Salzburg, Austria.
Organizational Affiliation: