Insulin Mimetic Peptide Disrupts the Primary Binding Site of the Insulin Receptor.
Lawrence, C.F., Margetts, M.B., Menting, J.G., Smith, N.A., Smith, B.J., Ward, C.W., Lawrence, M.C.(2016) J Biological Chem 291: 15473-15481
- PubMed: 27281820
- DOI: https://doi.org/10.1074/jbc.M116.732180
- Primary Citation of Related Structures:
5J3H - PubMed Abstract:
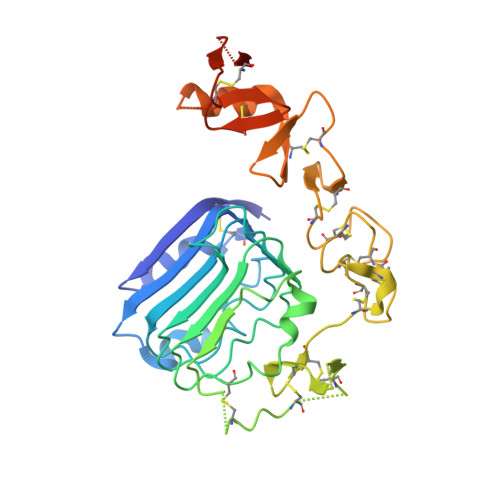
Sets of synthetic peptides that interact with the insulin receptor ectodomain have been discovered by phage display and reported in the literature. These peptides were grouped into three classes termed Site 1, Site 2, and Site 3 based on their mutual competition of binding to the receptor. Further refinement has yielded, in particular, a 36-residue Site 2-Site 1 fusion peptide, S519, that binds the insulin receptor with subnanomolar affinity and exhibits agonist activity in both lipogenesis and glucose uptake assays. Here, we report three-dimensional crystallographic detail of the interaction of the C-terminal, 16-residue Site 1 component (S519C16) of S519 with the first leucine-rich repeat domain (L1) of the insulin receptor. Our structure shows that S519C16 binds to the same site on the L1 surface as that occupied by a critical component of the primary binding site, namely the helical C-terminal segment of the insulin receptor α-chain (termed αCT). In particular, the two phenylalanine residues within the FYXWF motif of S519C16 are seen to engage the insulin receptor L1 domain surface in a fashion almost identical to the respective αCT residues Phe(701) and Phe(705) The structure provides a platform for the further development of peptidic and/or small molecule agents directed toward the insulin receptor and/or the type 1 insulin-like growth factor receptor.
- From the Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3052, Australia.
Organizational Affiliation: